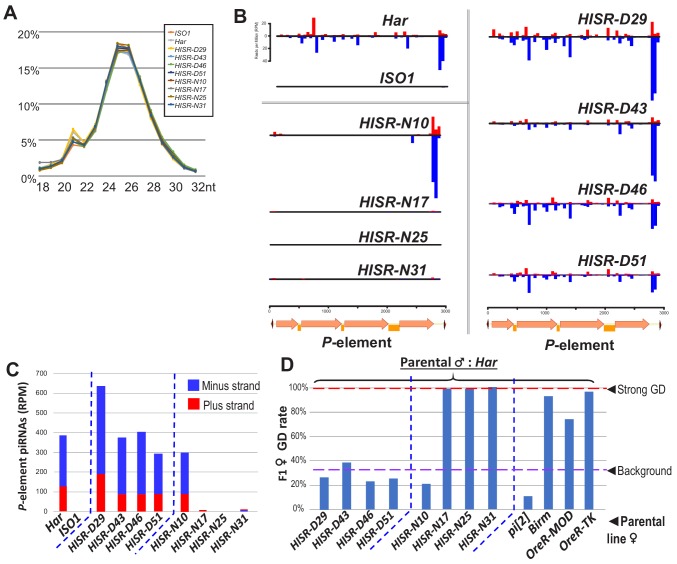
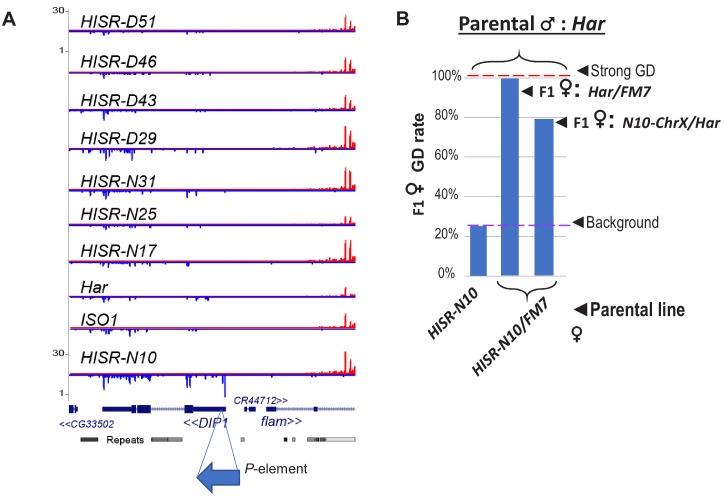
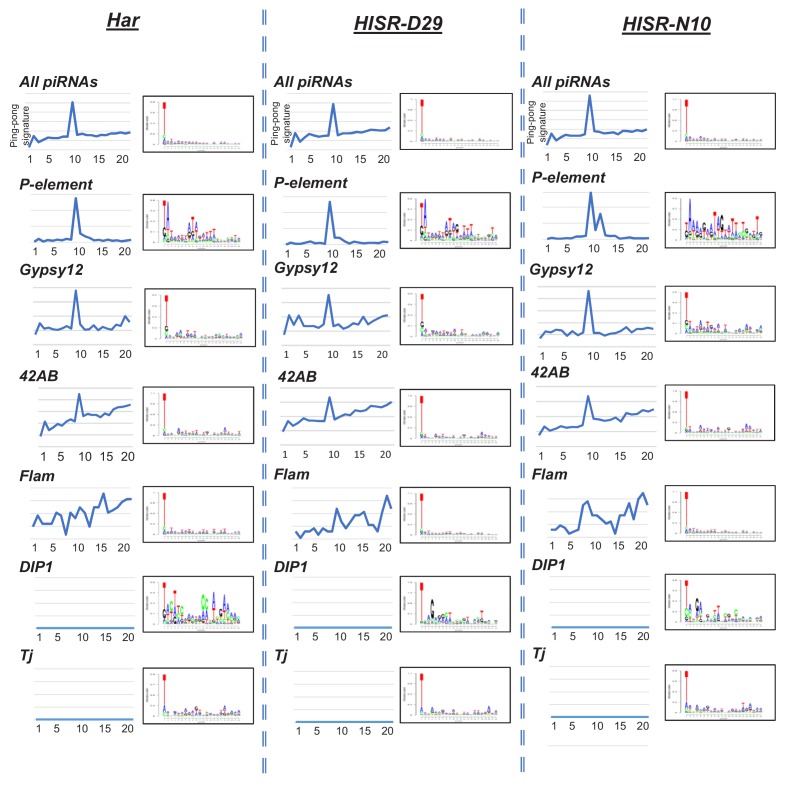
Figure 3. P-element directed piRNA production in HISR strains ovaries.
(A) Nearly identical size distributions of small RNAs from parental and HISR ovaries. (B) P-element piRNAs coverage plots and (C) quantitation of the P-element piRNAs mapping to plus and minus strands, in reads per million (RPM). (D) Assays for repression of P-element induced GD for HISR strains (N ≥ 100 females) are a good proxy for production of piRNAs silencing P-elements.