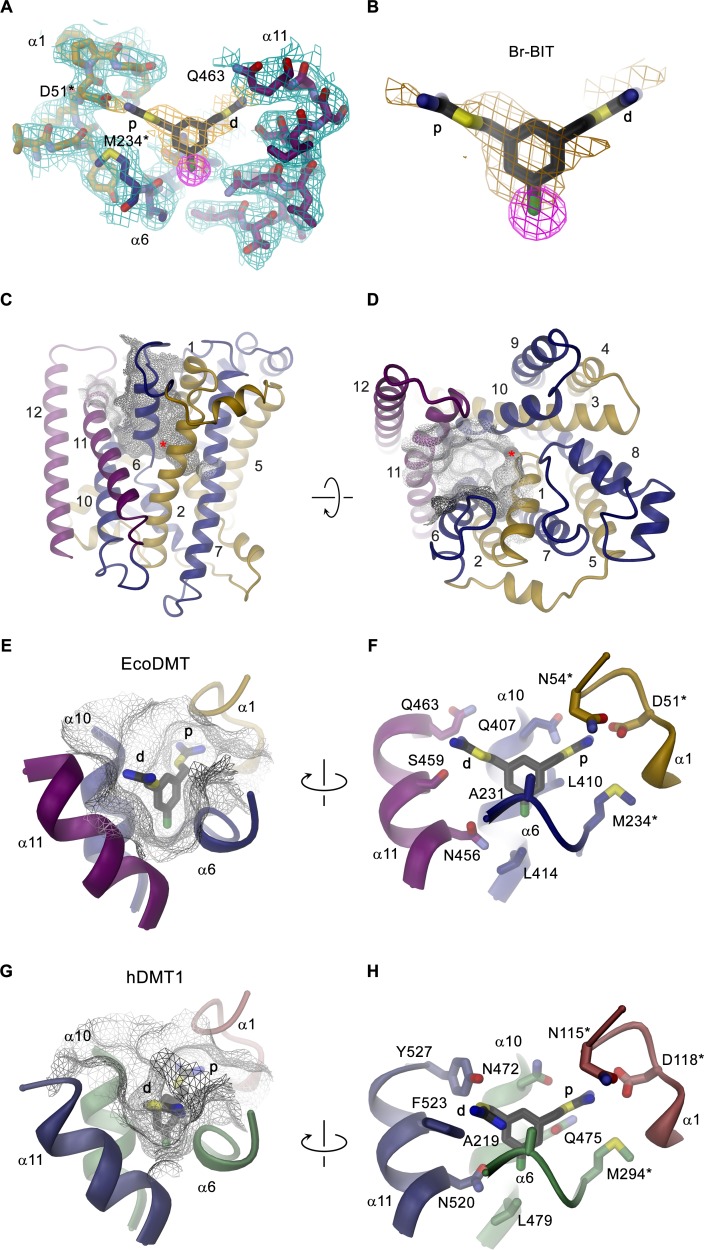
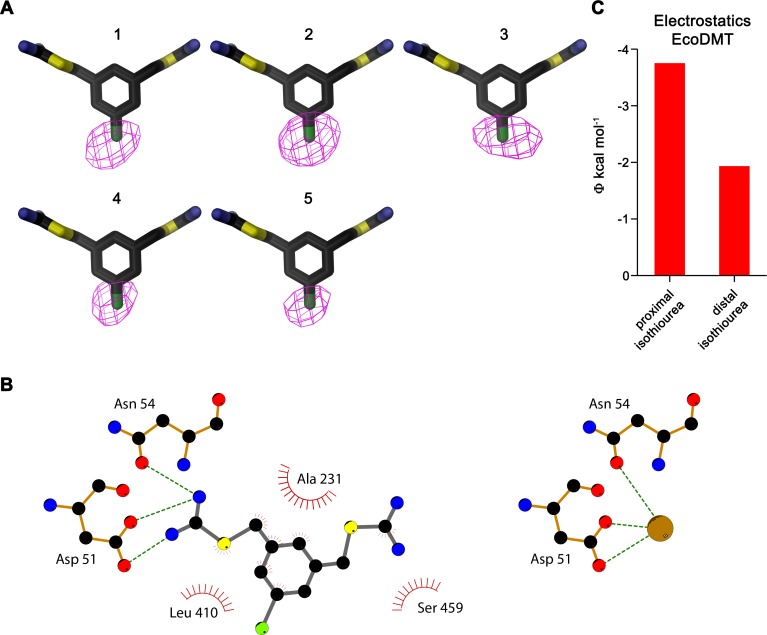
Figure 3. Structure of inhibitor complexes.
(A-B) Close-up views of the crystal structure of EcoDMT in complex with Br-BIT determined at 3.8 Å resolution viewed from within the membrane. The refined 2Fo-Fc electron density is shown as blue mesh. The position of Br-BIT is defined by the anomalous difference density of the Br-atom (shown as magenta mesh, contoured at 7σ) and by residual density in the 2Fo-Fc omit map (dark yellow mesh). C-D, Ribbon representations of the EcoDMT structure viewed from within the membrane (C) and from the extracellular side (D). The Mn2+ binding site is indicated with a red asterisk. The molecular surfaces are represented as gray meshes. The C-terminal sub-domain (α-helices 6–12) is shown in dark blue, α-helices 11 and 12 in magenta. (E) Position of Br-BIT in the binding pocket (gray mesh) of EcoDMT. (F) Detailed view of the residues in contact distance to Br-BIT. (G) Position of Br-BIT in the binding pocket (gray mesh) of a homology model of human DMT1. (H) Potential interactions of Br-BIT with the homology model of human DMT1. A-H, The proximal (p) isothiourea group is close to the metal ion coordinating residues (marked with a black asterisk) and the distal (d) isothiourea group is in proximity to α-helix 11.