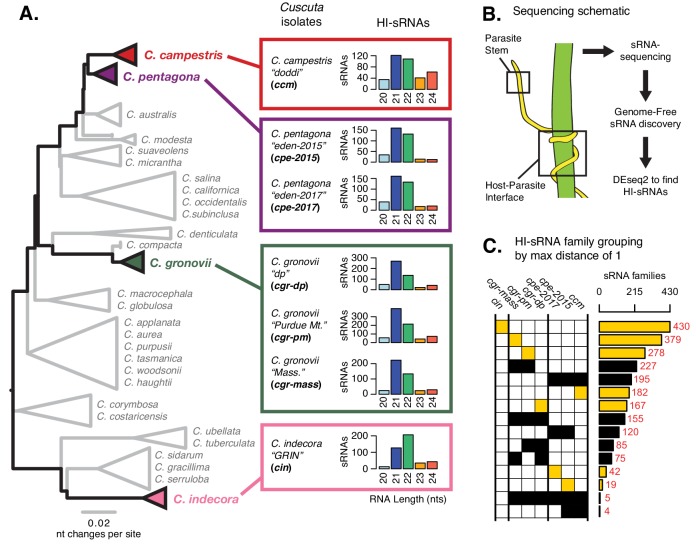
Figure 1. Haustorium-induced small RNAs (HI-sRNAs) are present in multiple Cuscuta species.
(A) Phylogeny of select Cuscuta species. Size distribution of HI-sRNAs for each sequenced isolate and acronyms are shown. (B) Sampling and sequencing schematic to discern HI-sRNAs. (C) HI-sRNA family counts and membership for each isolate, showing only the top 15 groups. Families were grouped strictly using a maximum edit distance of one nucleotide. Yellow indicates families present in a single isolate.