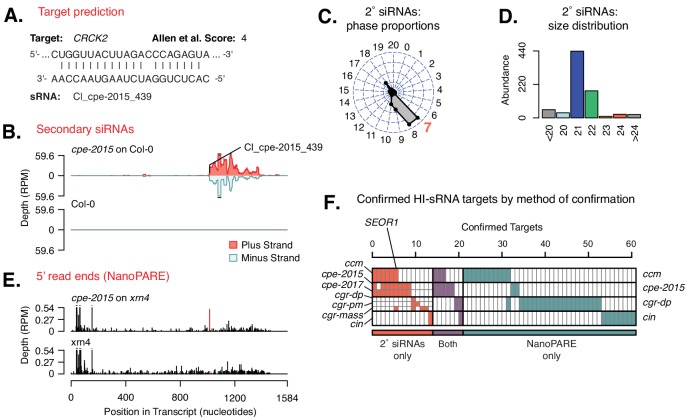
Figure 2. Host targets of Cuscuta HI-sRNAs.
(A) Modeled sRNA-target interaction for A. thaliana CRCK2. (B) Secondary siRNA accumulation from CRCK2. (C) Phasing analysis of secondary siRNAs from CRCK2. Expected phase for cut-site shown in red. (D) Size distribution of CRCK2 secondary siRNAs. (E) Frequency of 5’ ends from the CRCK2 mRNA, with the predicted HI-sRNA cut site shown in red. (F) Host mRNAs with confirmed targeting by a Cuscuta HI-sRNA. Full details in Figure 2—figure supplement 1 and Supplementary file 6.