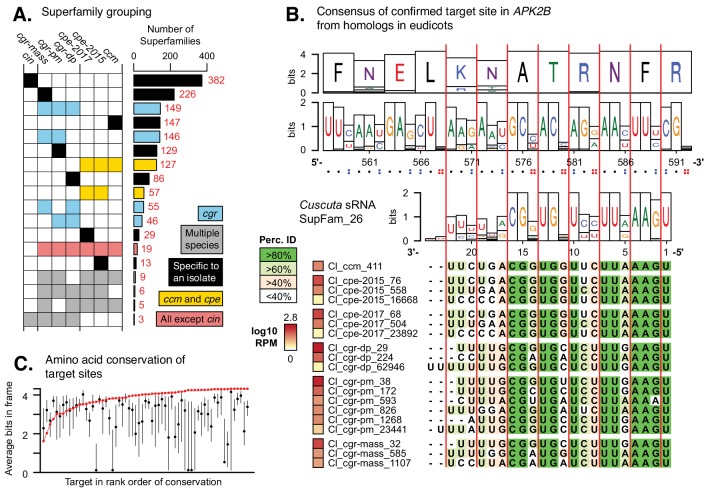
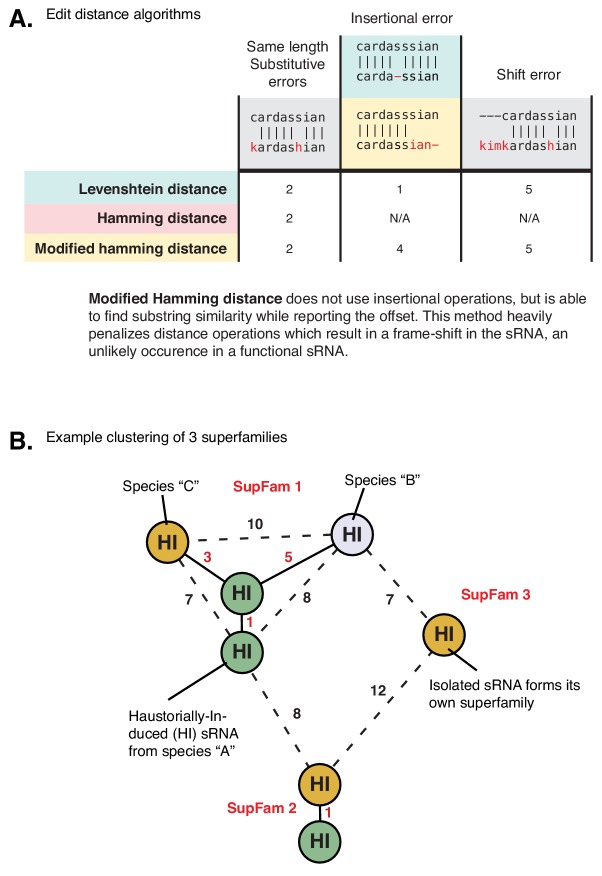
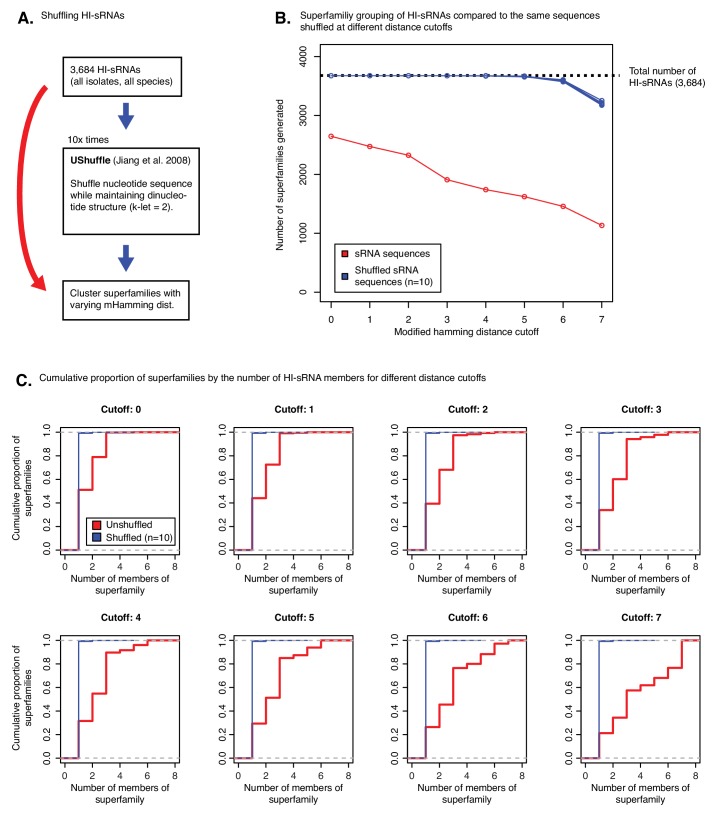
Figure 5. Cuscuta HI-sRNAs form superfamilies that co-vary with target sites across eudicots.
(A) sRNA superfamily count and membership for each Cuscuta isolate. Colors indicate general groupings of superfamilies. (B) An example HI-sRNA superfamily aligned to target sites from homologs in 36 eudicot genomes. Nucleotide and amino acid Shannon entropy from the alignments are shown as bits. Vertical red lines indicate the frame. Dots indicate the number of possible synonymous nucleotides at each codon. 17 additional examples in supplementary file 7. (C) Average conservation of target sites from homologs. Confirmed target site shown (red point), with all other possible sites shown by 25–75% quartiles (black line) and median (black point).