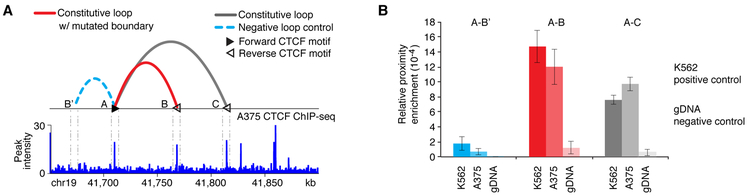
Figure 2. Validation of loop conformation around TGFB1.
(A) Top: Loop conformation around the TGFB1 associated insulator. Constitutive loop with the significantly mutated insulator (red), another constitutive loop (grey), and a negative control loop in chromosome conformation capture (3C) assays (light blue) are shown. CTCF motifs located at the two loop anchors are most likely to have forward-reverse orientation (faced arrows in black and white color). Bottom: CTCF ChIP–seq validation in A375 melanoma cells. The zoom-in shows the corresponding CTCF peaks at loop anchors A, B and C. (B) 3C assays confirm the existence of constitutive CTCF-CTCF loops between two pairs of CTCF anchors that are 60kb and 105 kb apart (red: loop A-B and grey: loop A-C) in A375 melanoma cells. To compensate for relative interaction efficiency based on linear distance, looping of anchor A with a random region upstream that was in closer linear distance than the downstream CTCF anchors (40kb), was also interrogated (light blue: loop A-B’) as a negative control loop. K562 leukemia cells were used as a positive control and genomic DNA that was purified, digested and ligated was used as a negative control. Relative enrichment of all samples was normalized over intra-fragment PCR. Data are represented as mean ± SD.
See also Figure S12.