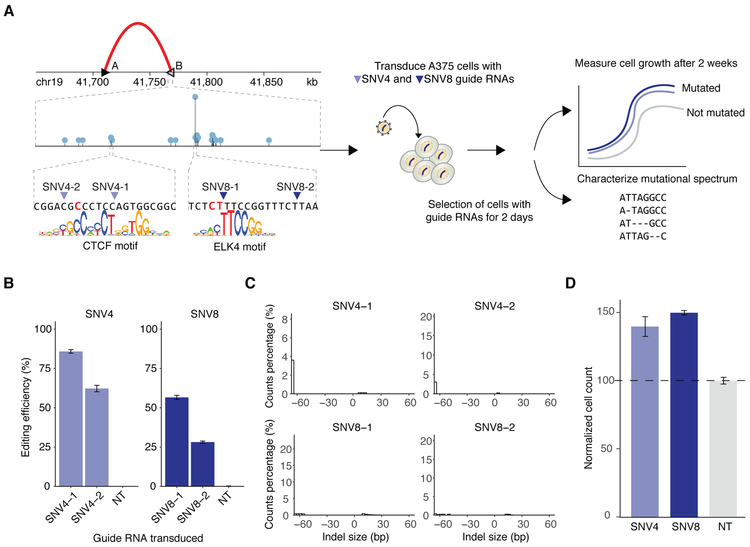
Figure 3. Functional validation of mutations in the TGFB1 associated insulator using CRISPR-Cas9 guide RNAs.
(A) Guide RNA design in the chr19 insulator region B. Needle plot shows mutations observed in B and zooms in on the sequence surrounding two of the most recurrent mutations in the region (SNV4 and SNV8). SNV4 is present in a CTCF motif and SNV8 is present in an ELK4 motif. Guide RNAs were designed to target 4 different positions (SNV4-1, SNV4-2, SNV8-1 and SNV8-2) surrounding SNV4 and SNV8. The guide RNAs are cloned into a lentiviral vector and transduced into human melanoma A375 cells. Cell proliferation is compared over a 2-week period between SNV4- and SNV8-edited cells to control cells transduced with non-targeting (NT) guide RNAs used as negative control. Deep sequencing readout characterizes the length of mutations in the selected cells. (B) Editing efficiency of each CRISPR guide RNA targeting SNV4 and SNV8. Data are represented as mean ± SEM. (C) Indel length distribution of each guide RNA that targets insulator region B. (D) Cell growth increases 40-50% in cells transduced with SNV4 and SNV8 guides. Data are represented as mean ± SEM. Cell count measurements are normalized to cells transduced with NT guide RNAs.
See also Figure S13.