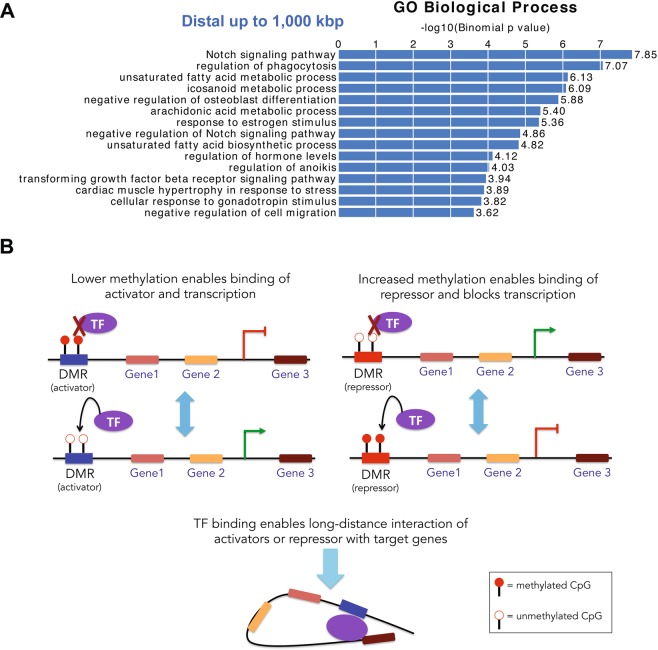
Figure 4.
DMRs distally located with respect to genes are enriched in biologically relevant pathways. (A) Pathways analysis using the Genomic Regions Enrichment of Annotations Tool (GREAT) that associates each gene with a ‘regulatory domain’ defined as a genomic region 5 kb upstream and 1 kb downstream from the TSS and an extension within 1 Mb up to the regulatory domain of the nearest upstream or downstream gene. (B) Schematic model depicting methylation-dependent modulation of TF binding to distal regulatory domains. Left diagram, hypermethylation of an activator inhibits, while hypomethylation of an activator facilitates TF binding, resulting in transcriptional repression or activation, respectively. Right diagram, particular TFs bind hypermethylated repressors, leading to transcriptional repression, while hypomethylation has the opposite effect on transcription. Bottom diagram, TF binding to a regulatory domain enables long-distance interactions with the target genes through 3D chromatin structural rearrangement.