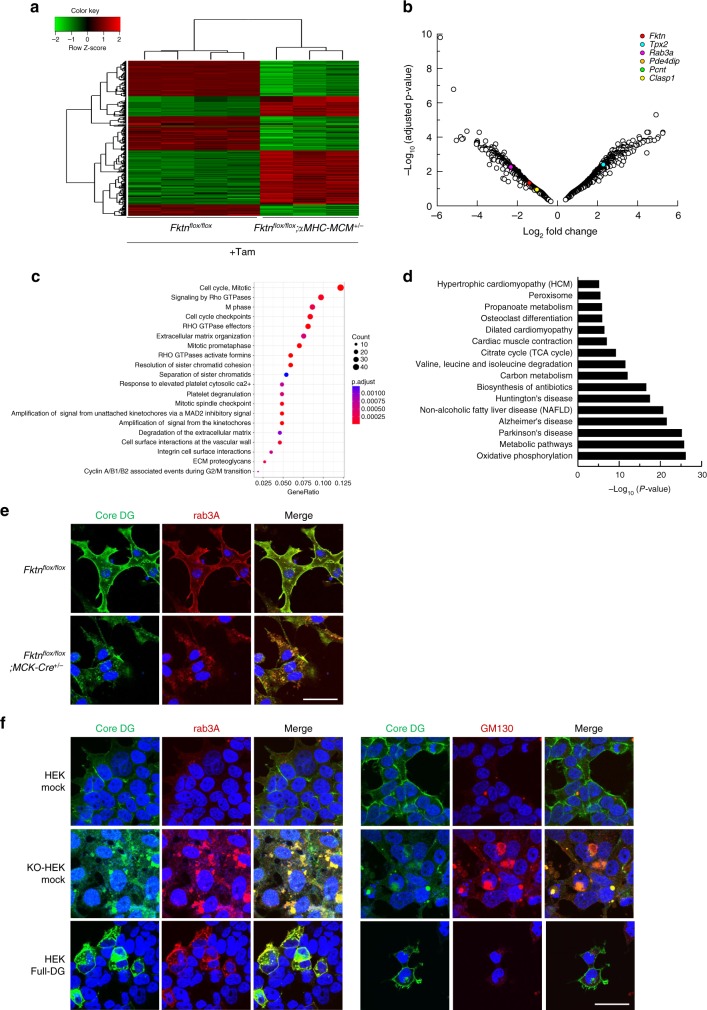
Fig. 10. Fktn elimination impacts Golgi-related genes and structure.
a Microarray analysis using floxed and αMHC-MCM-Fktn-cKO (Fktnflox/flox; αMHC-MCM+/−) hearts treated with tamoxifen for 4 d. Heat map displaying hierarchical clustering and expression levels of differentially expressed Golgi apparatus-related genes. b Volcano plot of upregulated or downregulated genes in tamoxifen-treated αMHC-MCM-Fktn-cKO (Fktnflox/flox; αMHC-MCM+/−) hearts. Dots indicating Pde4dip, Pcnt, and Clasp are overlapping. c Gene set enrichment analysis revealing enriched pathways and processes in tamoxifen-treated αMHC-MCM-Fktn-cKO hearts. d Significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) categories show differentially expressed gene pathways in tamoxifen-treated αMHC-MCM-Fktn-cKO hearts. e Representative immunofluorescence images of neonatal cardiomyocytes cultured with FCS (Core DG, green; Rab3a, red; DAPI, blue). Scale bar, 50 μm. f Representative immunofluorescence images of HEK, FKTN-KO HEK, and HEK expressing full-length DG (Core DG, green; Rab3a, red; DAPI, blue (left panels); Core DG, green; GM130, red; DAPI, blue (right panels)). Scale bar, 50 μm.