Figure 2.
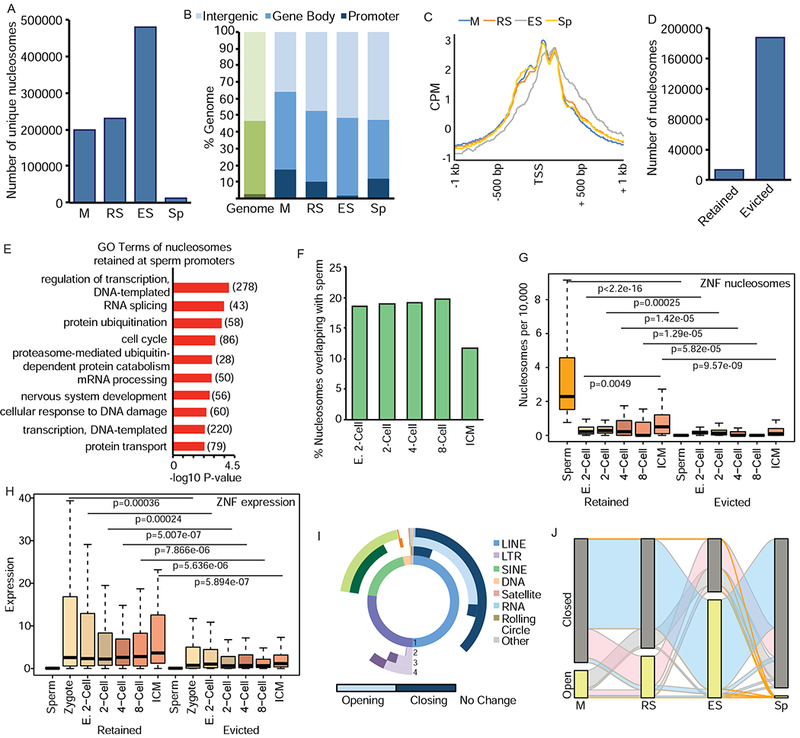
Nucleosome retention is mediated by dynamic changes in chromatin during spermiogenesis. A) Number of unique nucleosomes in germ cells following partitioning of ATAC-seq (180-247bp) and analysis by DANPOS. B) Genomic compartment analysis of nucleosome location in germ cells (blue) compared to genome (green). C) Meta-analysis of nucleosome position at TSS of genes with accessible nucleosomes. CPM=count per million reads. D) Number of nucleosomes evicted or retained in Sp previously present in M stage. E) Top enriched GO terms for retained Sp nucleosomes. Number of genes in parentheses. F) Percentage of nucleosomes retained in Sp overlapping with nucleosomes in pre-implantation embryos. E. 2-cell: early 2-cell, ICM: inner cell mass. G) Comparison of nucleosomes retained (left) or evicted (right) at ZNFs in Sp and pre-implantation embryos. H) Comparison of RNA expression at retained (left) or evicted (right) ZNFs in gametes and pre-implantation embryos. I) Donut plot depicting changes in REs from total ATAC-seq signal. (1) represents proportion of nucleotides for each class of REs in genome. (2) M to RS transition (3) RS to ES (4) ES to Sp. Light shades represent increased enrichment of RE, dark shades represent depletion. Lack of color (white) represents no change. J) Alluvial plot of nucleosomes located in open (detected by DANPOS, yellow bar) or closed (not detected, grey) chromatin compartments. Orange ribbons depict nucleosomes that are ultimately retained (present) in Sp, blue are present in ES but absent in Sp, pink are present in RS, but absent in ES and Sp, grey ribbons present only in M. See also Figure S2.
