Figure 6.
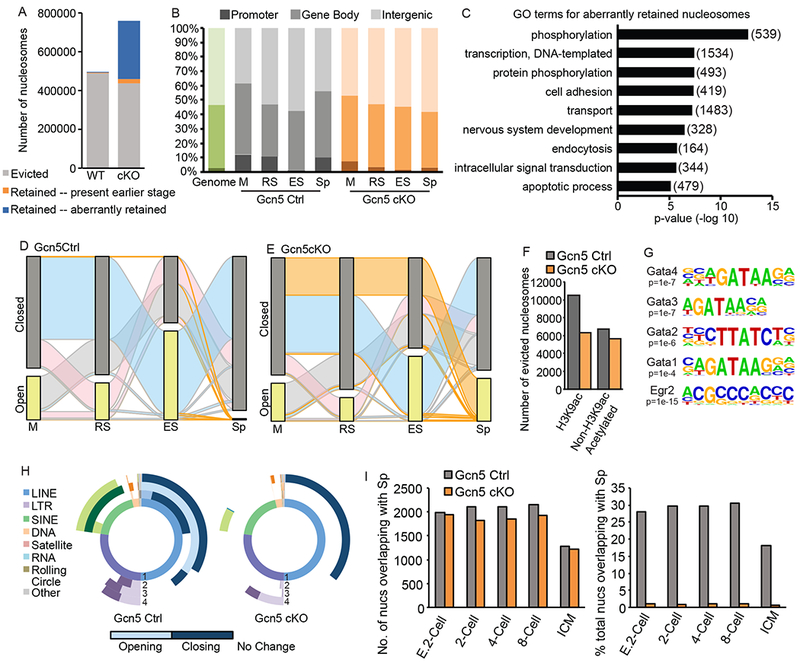
Aberrant nucleosome retention in Gcn5cKO sperm. A) Number of evicted (grey), retained from any previous stage (orange), or newly accessible (blue) nucleosomes in Gcn5Ctrl or Gcn5cKO Sp. B) Compartment analysis of nucleosome location based on percent of total nucleosomes (green bar, genomic background). C) Top enriched GO terms for aberrantly retained Gcn5cKO nucleosomes (number of genes in parentheses). Alluvial plot of nucleosomes in open (detected in specific cell type, yellow bar) or closed (not detected, grey) nucleosomes in (D) Gcn5Ctrl or (E) Gcn5cKO. Ribbon colors as described above. F) Number of evicted nucleosome dense regions with H3K9ac (defined by ChIP-seq, Bryant et. al., 2015) in RS. G) Enriched TF binding motifs present in newly open subnucleosomal regions in Gcn5cKO Sp. H) Donut plot depicting changes in REs from total ATAC-seq signal for Gcn5Ctrl (top) or Gcn5cKO (bottom). Plots as described above. F) Percentage of nucleosomes retained in Sp overlapping with nucleosomes in pre-implantation embryos. E. 2-cell: early 2-cell, ICM: inner cell mass. See also Figure S5.
