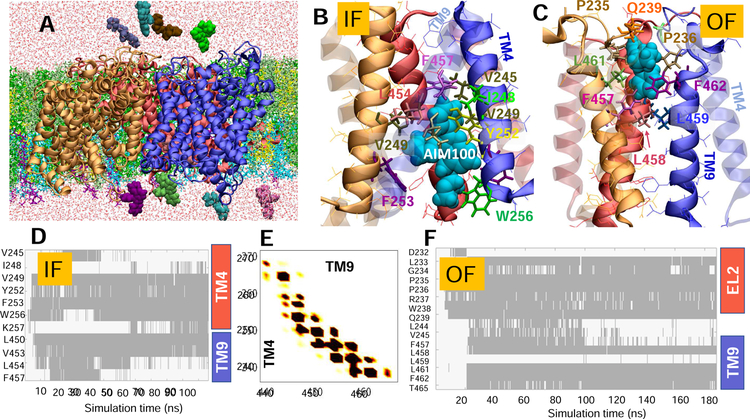
Fig. 2. Simulations of AIM-100 binding to DAT trimer-W238 highlight the involvement of EL2b, TM4 and TM9 in stabilizing the trimer and coordinating the drug.
(A) Typical set up for MD simulations. The trimer-W238 (protomers colored blue, red and orange) was embedded into neuronal lipids (licorice) containing POPE (green), POPC (lime), cholesterol (yellow), POPI (cyan), and PIP2 (purple) molecules. AIM-100 molecules (vDW, various colors) were initially distributed in both the EC and IC regions. Multiple binding events were observed. (B) AIM-100 (cyan, vdW) bound to the IC pore of IF trimer, stabilized by TM4 (solid) and TM9 (transparent); and (C) AIM-100 bound to the EC pore of the OF trimer, lined by TM9 (solid) and EL2. (D) Time evolution of contacts between IF trimer-W238 and AIM-100. Contacts are indicated by gray-shaded areas. (E) Interprotomer contacts between TM4 and TM9, observed during the simulations of the IF trimer (see also Fig S2). (F) Same as D for OF trimer shown in C (see also Fig S3). In D-F, contacts refer to atom-atom distances closer than 3.5 Å.