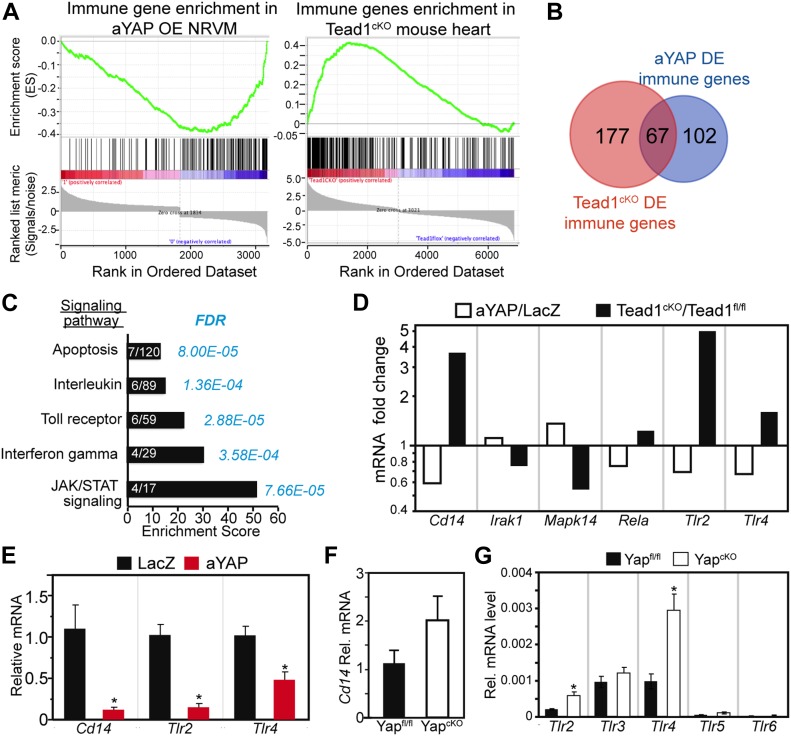
Figure 5. YAP/TEAD1 regulates innate immune gene expression.
(A) GSEA. DE (DE) genes from YAP gain-of-function or Tead1 loss-of-function data set were analyzed against an immunology gene list. YAP OE NRVMs with YAP gene overexpressed. Tead1cKO, heart-specific Tead1 knockout. Immune genes were enriched in the down-regulated genes in YAP OE NRVM and in the up-regulated genes of Tead1cKO heart. (B) Venn diagram of three different gene list sets: immunology genes, DE genes in the Tead1cKO data set, and DE genes in the YAP OE NRVM data set. In these three gene list sets, 67 common genes were identified. (C) Signaling pathway GO term analysis. The list of 67 genes identified in (B) was used for GO term signaling pathway enrichment analysis (http://geneontology.org). PANTHER overrepresentation test tool was used to analyze the enriched signaling pathways. Bar graph was plotted based on enrichment score. Gene number enriched in related pathways was labeled on each bar. Statistical significance was shown by false discovery rate value. (D) mRNA of Cd14 and Toll receptors and in YapcKO mouse heart. The expression values of different Tlr genes were normalized to Gapdh. RNA was collected from 1 mo old Myh6:Cre; Yapfl/fl (YapcKO) mouse heart. N = 4. *P < 0.05. (E) qRT-PCR measurement of Cd14, Tlr2, and Tlr4 in the NRVMs. NRVMs were treated with indicated virus for 48 h in the absence of serum. N = 4. t test: *P < 0.05. (F, G) mRNA of Cd14 (F) or Tlr genes (G) in 1 mo old Myh6:Cre; Yapfl/fl (YapcKO) mouse hearts. The expression values of different Tlr genes were normalized to GAPDH. N = 4. t test: *P < 0.05.