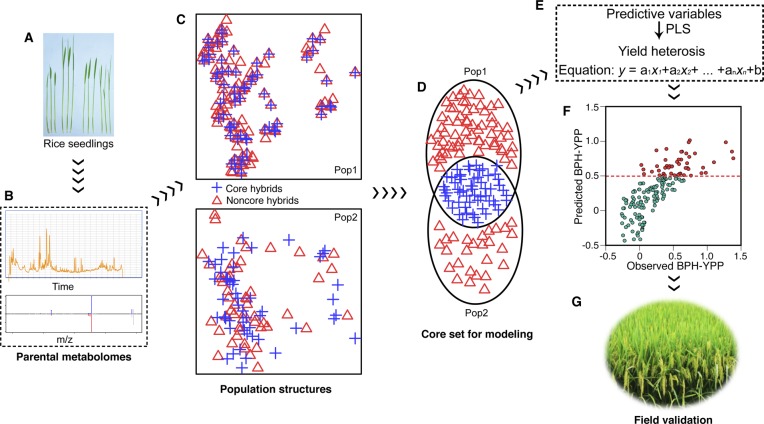
Figure S12. Schematic diagram of precision breeding 1.0.
(A, B, C, D, E, F, G) Two hybrid populations were selected as part of the untargeted metabolome-based prediction strategy. Seedlings of parents from both populations (Pop1 and Pop2) were grown under uniform conditions (A). After performing LC-MS analysis of ∼15-d-old seedlings, parental metabolite data were obtained (B), and the mean relative abundances of all detected analytes between every pair of parents were calculated as predictive variables for corresponding hybrids. PCA of hybrids from both populations with all predictive variables was performed to dissect population structure (C). Core hybrids selected from both populations (marked in blue) according to principal component 1 scores were used as a training set. The hybrids in the training set were field phenotyped under corresponding conditions. The PLS regression analysis was used to find connections between predictive variables and hybrid performance for the training set (D). The average VIP values of all latent factors were used to remove low-contribution or unrelated predictive analytes. Next, a predictive model was established according to the parameter of each variable and the constant (E). Finally, the phenotypes of candidate hybrids were predicted (F), and hybrids with predicted high degree of heterosis were field phenotyped to determine the elite combinations (G).