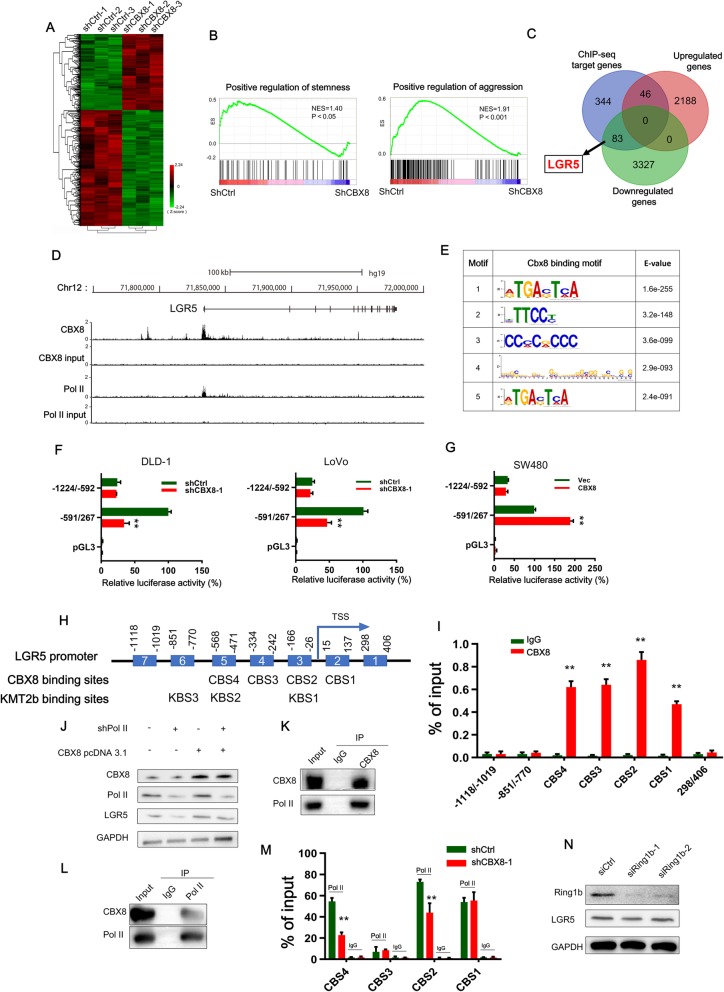
Fig. 3.
CBX8 regulates LGR5 transcription by interacting with Pol II in a noncanonical manner. a, The heatmap of the differentially expressed genes after CBX8 knockdown. b, GSEA showed that genes differentially expressed in response to CBX8 knockdown were enriched in gene sets significantly related to stemness and cancer aggression. c, The Venn diagram showing the numbers of overlapping genes between the set of genes differentially expressed by CBX8 knockdown and the set of target genes identified by ChIP-seq. d, Overview of the LGR5 promoter region with ChIP-seq data for CBX8 and Pol II in DLD-1 cells. e, The top five predicted CBX8-binding elements were obtained by de novo motif analysis using MEME software. f, Luciferase reporter genes driven by the − 1224/− 592 or − 591/267 fragments of the LGR5 promoter region were cotransfected with shCtrl or shCBX8 into DLD-1 and LoVo cells, and luciferase activity was measured after 48 h. The relative luciferase activity value in cells cotransfected with pRL-TK (− 591/267) and shCtrl was set to 100%. g, Luciferase reporter genes driven by the − 1224/− 592 or − 591/267 fragments of the LGR5 promoter region were cotransfected with the empty vector (vec) or the CBX8 overexpression pcDNA3.1 plasmid into SW480 cells, and luciferase activity was measured after 48 h. h, A schematic of the seven LGR5 promoter regions (1–7) analyzed for CBX8 binding affinity (above). Schematic representation of predicted CBX8 and KMT2b binding sites (below). i, ChIP-qPCR analysis was used to determine the binding affinity of CBX8 to seven LGR5 promoter regions in DLD-1 cells, showing that CBX8 bound to the CBS1-CBS4 regions in the LGR5 promoter. ChIP-qPCR with IgG was performed as the control. j, Western blot analysis of LGR5 expression in Pol II knockdown DLD-1 cells with or without overexpressing CBX8. k-l, CoIP was used to show the interaction between the CBX8 and Pol II proteins in DLD-1 cells. m, ChIP-qPCR analysis was used to assess the binding affinity of Pol II to the CBS1-CBS4 regions after CBX8 knockdown in DLD-1 cells. n, Western blot was used to determine the expression of Ring1b and LGR5 after silencing of Ring1b in DLD-1 cells. Data are shown as the mean ± SD of three replicates (*, P < 0.05; **, P < 0.01)