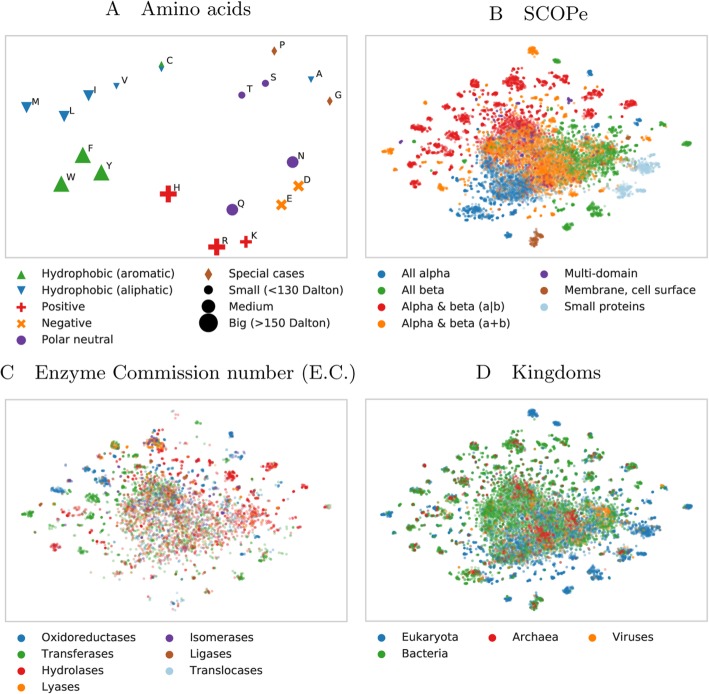
Fig. 3.
Modeling aspects of the language of life. 2D t-SNE projections of unsupervised SeqVec embeddings highlight different realities of proteins and their constituent parts, amino acids. Panels B to D are based on the same data set (Structural Classification of Proteins – extended (SCOPe) 2.07, redundancy reduced at 40%). For these plots, only subsets of SCOPe containing proteins with the annotation of interest (enzymatic activity C and kingdom D) may be displayed. Panel A: the embedding space confirms: the 20 standard amino acids are clustered according to their biochemical and biophysical properties, i.e. hydrophobicity, charge or size. The unique role of Cysteine (C, mostly hydrophobic and polar) is conserved. Panel B: SeqVec embeddings capture structural information as annotated in the main classes in SCOPe without ever having been explicitly trained on structural features. Panel C: many small, local clusters share function as given by the main classes in the Enzyme Commission Number (E.C.). Panel D: similarly, small, local clusters represent different kingdoms of life