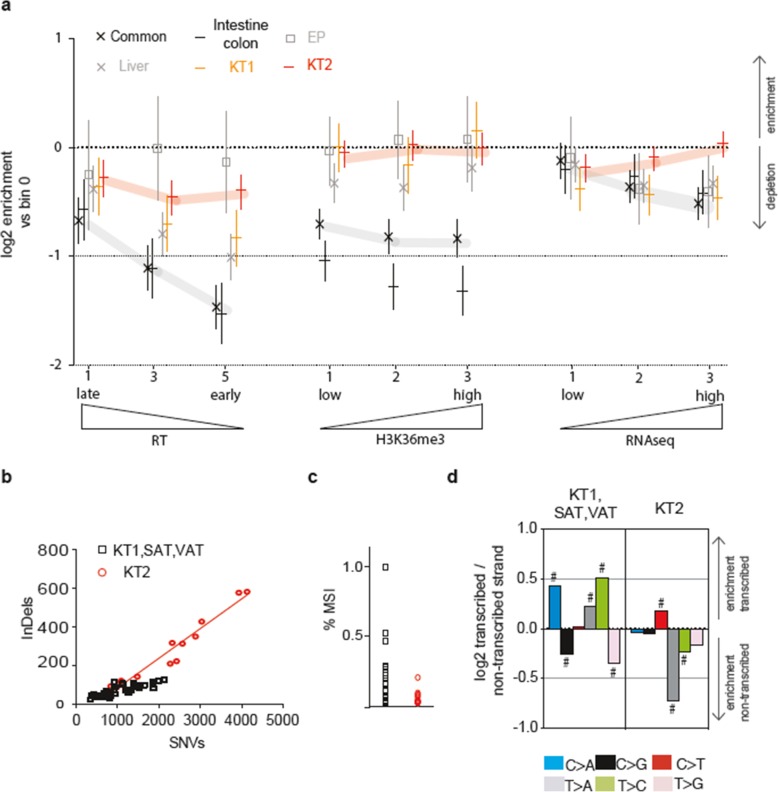
Fig. 6.
Mutation enrichment in specific genomic regions provides information on DNA repair efficiency and mutagen exposure in different cell types. a Enrichment/depletion of mutations in specific genomic regions. The genomes were divided in multiple sectors (bins) according to decreasing DNA replication time (RT, bins 0 to 5. For clarity, only bins 1, 3, and 5 are shown), increasing abundance of the histone mark H3K36me3 (bins 0–3), and increasing transcriptional levels (RNA-seq, bins 0–3). The relative abundance of mutations in each bin vs bin 0 for every tissue (EP, liver, KT1, KT2) or tissue group (common progenitors: SAT, VAT, SkM, blood; intestine-colon) is estimated as the coefficient in negative binomial regression (expressed as log2), where error bars show its 95% C.I. b Linear regression of SNVs and InDels per genome in the KT2 vs KT1-SAT-VAT group. c Percentage of sites subjected to microsatellite instability (MSI) in each genome of either the KT2 or the KT1-SAT-VAT group. d Enrichment of the six classes of substitution types in either transcribed or non-transcribed strand of genes. The log2 ratio of the number of observed and expected point mutations indicates the effect size of the enrichment in the transcribed (upper) or non-transcribed (lower) strand. #P < 0.05, one-sided binomial test