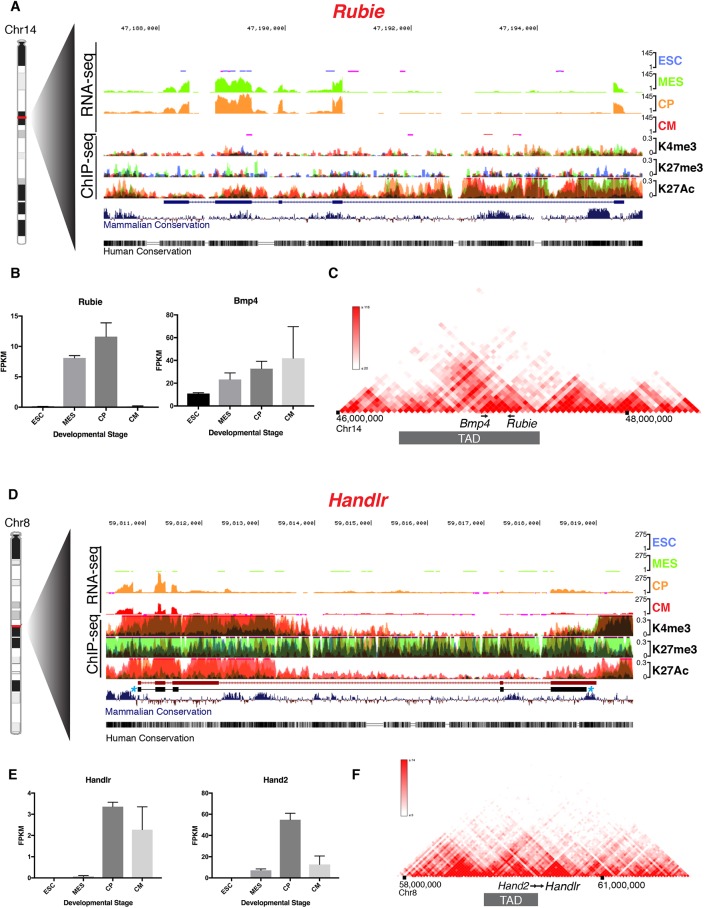
Fig. 2.
Genomic characterization of Rubie and Handlr in vitro. (A) UCSC Genome Browser tracks of Rubie RNA-seq and overlaid histone H3 ChIP-seq at ESC, MES, CP and CM stages of in vitro differentiation; RefSeq annotation in blue. (B) Quantified expression of Rubie and Bmp4 at each differentiation stage. (C) 3D Genome Browser Hi-C heatmap of chromosome interactions around Bmp4 and Rubie loci. (D) UCSC Genome Browser tracks of Handlr RNA-seq and overlaid histone H3 ChIP-seq at ESC, MES, CP and CM stages of in vitro differentiation. Ensembl annotation in red; observed exon structure of predominant Handlr transcript in black with blue asterisks. (E) Quantified expression of Handlr and Hand2 at each differentiation stage. (F) 3D Genome Browser Hi-C heatmap of chromosome interactions around Handlr and Hand2 loci. ESC, embryonic stem cell; MES, mesoderm; CP, cardiac progenitor; CM, cardiomyocyte; blue, ESC; green, MES; orange, CP; red, CM; K4me3, histone H3 lysine 4 trimethylation; K27me3, histone H3 lysine 27 trimethylation; K27Ac, histone H3 lysine 27 acetylation; TAD, topologically associated domain. Data are mean±s.e.m. in B and E.