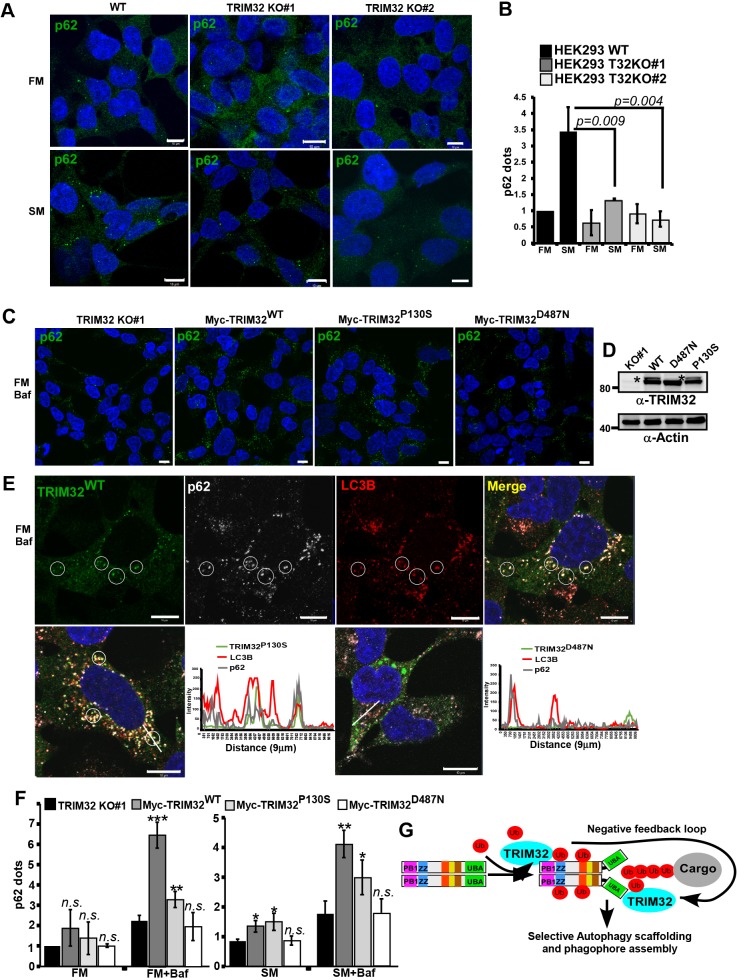
Fig. 6.
TRIM32 enhances the formation and turnover of p62 dots. (A) Immunostaining of endogenous p62 in HEK293 FlpIn cells and the two TRIM32 KO clones under normal (FM) and starvation (SM) conditions (HBSS 2 h). Scale bars: 10 µm. (B) Quantification of p62 dots in the two TRIM32 KO clones and the HEK293 FlpIn wild type cell line under normal (FM) and starvation (SM) (HBSS 2 h) conditions using the Volocity software (PerkinElmer). Each graph represents the mean±s.d. of the relative number of p62 dots per cell (normalized to the value for FM in wild-type cells). The results are from three independent experiments each including ∼100 cells per condition. P-values were obtained using Student's t-test. (C) Immunostaining of endogenous p62 in the TRIM32 KO (#1) cell line and TRIM32 KO (#1) cells reconstituted with stable expression of Myc–TRIM32WT, Myc–TRIM32P130S or Myc–TRIM32D487N as indicated. The cells are grown in FM with BafA1 (2 µM, 7 h). Scale bars: 10 µm. (D) Western blot showing the expression of Myc–TRIM32, Myc–TRIM32P130S and Myc–TRIM32D487N in the reconstituted HEK293 TRIM32-KO cell lines, grown in FM. * indicates mono-ubiquitylated TRIM32. (E) Upper panels show immunostaining of Myc–TRIM32WT, p62 and LC3B in the reconstituted HEK293 TRIM32-KO cells grown in normal medium supplemented with BafA1 (2 µM) for 7 h. The circles exemplify colocalization of all three proteins in BafA1 dots. The lower panels show merged images of similar immunostainings in the TRIM32-KO cell line reconstituted with Myc–TRIM32P130S or Myc–TRIM32D487N. Images were obtained using a ZEISS780 confocal laser scanning microscope, and the colocalization line plots to the right obtained using the ZEN software. Scale bars: 10 µm. (F) Quantification of p62 dots in the cell lines represented in C, under normal (FM) and normal plus BafA1 (7 h) conditions, and starvation (SM) (HBSS 4 h) and starvation plus BafA1 conditions. The graphs represent the mean±s.d. of the relative number of p62 dots per cell (normalized to the value for FM in TRIM32-KO cells) quantified using Volocity software (PerkinElmer). The results are from three independent experiments each including n>100 cells per condition. *P<0.05, **P<0.005, ***P<0.0005; n.s., not significant (Student's t-test). (G) A model depicting the roles of TRIM32 both as a p62 activator and a p62 substrate in selective autophagy. ZZ, zinc finger; UBA, ubiquitin-binding domain; Ub, Ubiquitin.