Fig. 1.
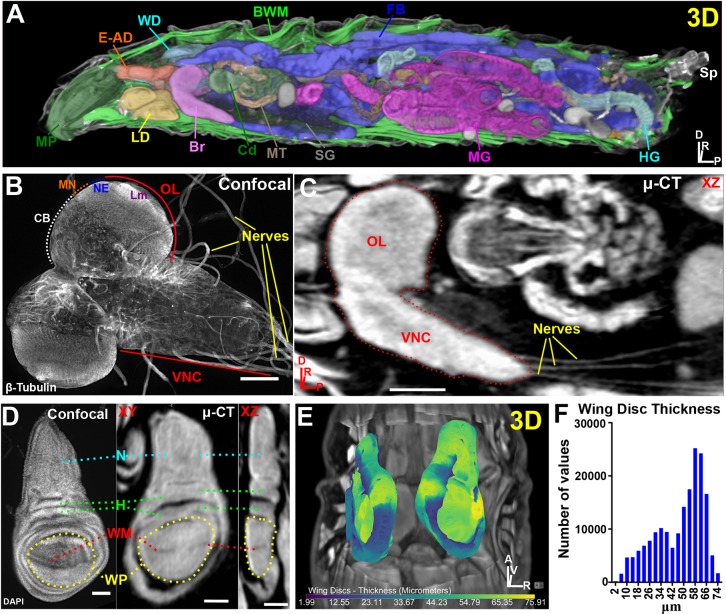
µ-CT reveals all major organs in Drosophila melanogaster larvae. (A) 3D display of a larva optically sliced along the xz axis showing the mouthparts (MP), eye-antennal discs (E-AD), wing discs (WD), body wall muscles (BWM), fat body (FB), spiracles (Sp), hindgut (HG), midgut (MG), salivary glands (SG), Malpighian tubules (MT), cardia (Cd), brain (Br) and leg discs (LD). (B) Confocal image of explanted larval brain labeled with β-tubulin to visualize nerves. Ventral nerve cord (VNC), optic lobe (OL), central brain (CB), medulla neuroblasts (MN), neuroepithelium (NE) and lamina (Lm) neuropil are denoted. (C) µ-CT of larval brain (dotted outline) viewed along the xz axis. Note extension of nerves to target organs from the VNC. (D) Confocal image of DAPI-stained wing disc (left) and a µ-CT-imaged wing disc (right). Notum (N), hinge (H), wing margin (WM) and wing pouch (WP) are labeled; also note epithelial folds in the xz view. (E) 3D representation of wing disc position within the body cavity. Discs are colored by thickness (µm) (see Materials and Methods). (F) Histogram of wing disc thickness values. Axis denotes origin; A, anterior; D, dorsal; P, posterior; R, right; V, ventral. Scale bars: 100 µm (B,C); 50 µm (D). Stained with 0.1 N iodine and scanned in slow mode at an image pixel size of 2 µm.