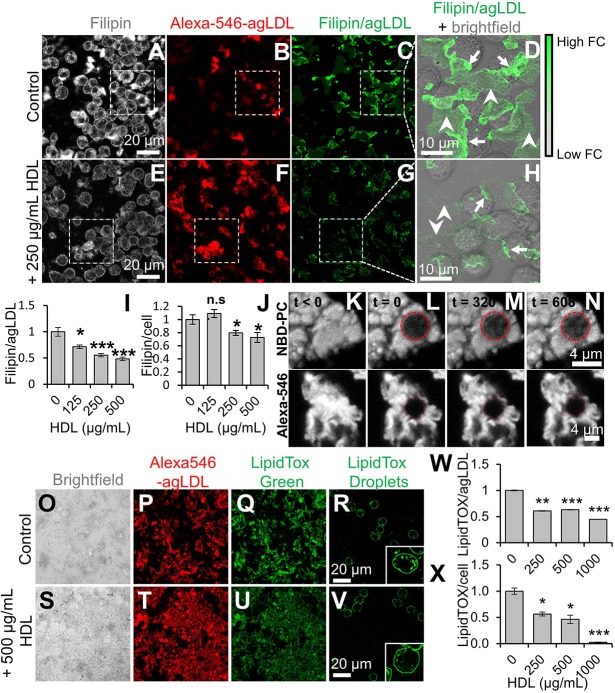
Fig. 2.
HDL reduces macrophage FC uptake from agLDL and foam cell formation. (A–H) J774 macrophages were incubated with cross-linked Alexa546–agLDL for 4 h in absence (A–C) or presence (E–G) of 250 µg/ml HDL. Cells were fixed, FC stained with filipin, and cells analyzed by confocal microscopy. Filipin and agLDL images (C,G) show FC intensity relative to agLDL fluorescence. (D,H) Filipin and agLDL from areas highlighted by the white dashes boxes in C and G overlaid on corresponding bright-field images. Arrows show agLDL in contact with cells, where FC is enriched due to catabolism of agLDL. Arrowheads show agLDL not directly in contact with cells. Images taken as described in A–H for cells in the presence of the indicated amount of HDL were used to quantify (I) the ratio of FC to agLDL and (J) the amount of free cholesterol relative to that in control cells (set at 1) for at least 10 fields containing >100 cells. (K–N) agLDL was labeled with NBD-PC and (K) observed prior to photobleaching. (L) agLDL was photobleached at t=0 in the area indicated by the red dashed circle. Fluorescence recovery after photobleaching within the photobleached area was observed after (M) 320 s and (N) 608 s. (O–V) J774 macrophages were incubated with Alexa546–agLDL stabilized by a collagen matrix for 24 h at 37°C in the presence of 0, 250, 500 or 1000 µg/ml HDL. Cells were fixed, and cholesteryl esters were stained with LipidTox Green and analyzed by confocal microscopy. Shown are bright-field, Alexa546–agLDL, total LipidTox Green and cell-associated LipidTox Green images of cells incubated with 0 (O–R) and 500 (S–V) µg/ml HDL. Cell-associated LipidTox Green images were obtained after background subtraction of the agLDL-associated LipidTox Green signal. Insets show a representative cell from the same field. Images were used to quantify agLDL-associated LipidTox fluorescence (W) and cell-associated LipidTox fluorescence (X) for at least 10 fields containing >100 cells relative to that in control cells (set at 1). Data were compiled from at least three independent experiments. Error bars show s.e.m. *P≤0.05; **P≤0.01; ***P≤0.001; n.s. not statistically significant.