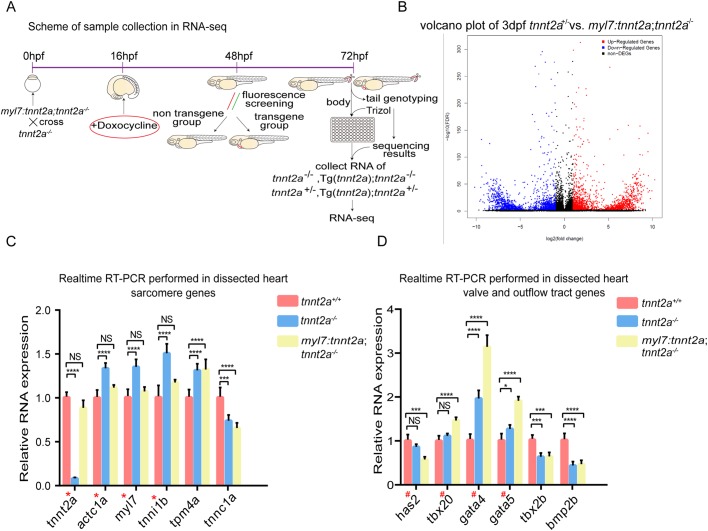
Fig. 3.
Gene expression in OFT is deregulated in tnnt2a mutant zebrafish with myocardial tnnt2a supplementation. (A) Workflow of sample collections for whole-fish RNA-seq of four groups: tnnt2+/−, tnnt2a−/− Tg(myl7:tnnt2a); tnnt2a−/− and Tg(myl7:tnnt2a); tnnt2a+/−. (B) Volcano plot of differentially expressed genes in tnnt2a+/− versus Tg(myl7:tnnt2a); tnnt2a−/−. (C,D) Sarcomere- and OFT-relevant gene expression in dissected heart of tnnt2a+/+, tnnt2a−/−, and Tg(myl7:tnnt2a); tnnt2a−/− after DOX induction at 16 hpf. *P<0.05, ***P<0.001, ****P<0.0001, NS, not significant; n=30 per group. Statistical differences were evaluated using two-way ANOVA. Red asterisks, sarcomere genes in Tg(myl7:tnnt2a); tnnt2a−/− can almost return to wild-type level after DOX induction; red hashes, OFT-relevant genes in Tg(myl7:tnnt2a); tnnt2a−/− are more deviated from wild-type level after DOX induction compared with tnnt2a−/−.