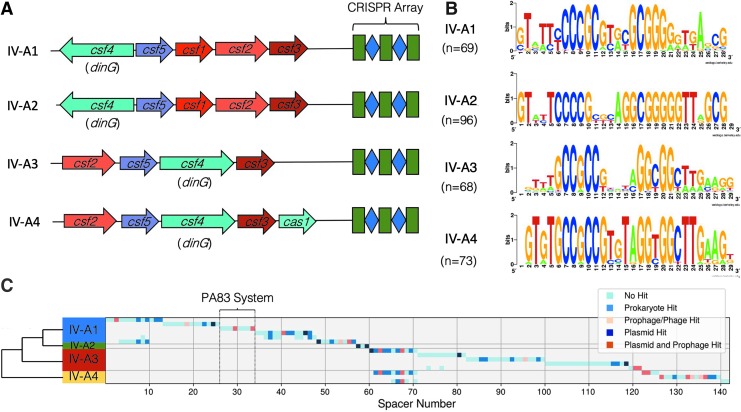
FIG. 1.
Type IV-A CRISPR-Cas systems found in Pseudomonas aeruginosa. (A) Classification of type IV-A CRISPR-Cas systems found in P. aeruginosa. Conserved architectures of these four variants are observed. All four variants include csf2, csf3, csf4/dinG, and csf5/cas6. Despite the similar architecture between type IV-A1 and type-IV-A2, they represent two separate groups based on differences in direct repeat sequences and amino acid divergence (Supplementary Fig. S1). (B) Consensus sequences for direct repeats found in type IV-A CRISPR arrays in P. aeruginosa. Palindromic regions are in the center of the repeat. (C) Table of spacers found in each system. Systems are grouped by variant, with group 2 consolidated, as all their CRISPR arrays are identical. Spacers are colored based on mapping results according to the legend. A dendrogram was generated based on representative csf2 sequences to illustrate the relationship between variants.