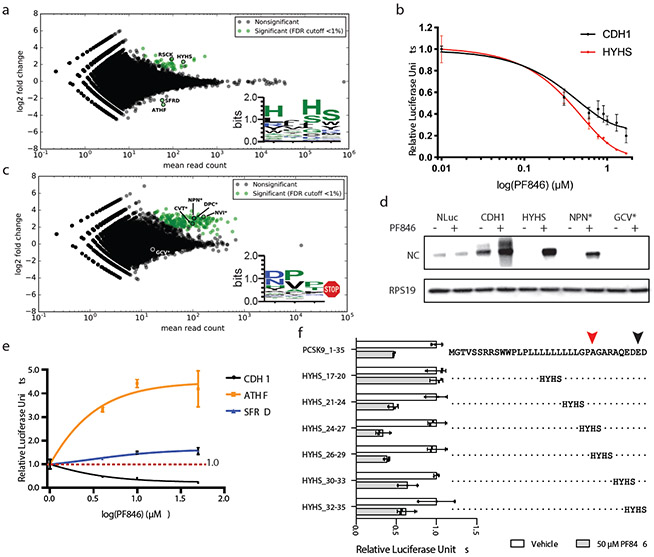
Fig. 5 ∣. Selections of PF846-dependent stalling sequences from randomized mRNA libraries.
a, MA plot (data with M (log ratio) and A (mean average) scales) of sequences enriched for PF846-induced translation elongation stalling, compared to translation reactions in the absence of PF846. Sequences were enriched from the mRNA library of CDH1-derived nascent chains with four amino acid positions randomized near the predicted stall site in the PTC. Log2-fold enrichment of sequences is plotted against total read count, for experiments carried out in duplicate. Green, sequences enriched with an adjusted P-value < 0.01. Inset, consensus sequence logo 38 for enriched sequences. b, In vitro translation assays with luciferase reporters containing the WT CDH1 stalling sequence or with the HYHS motif near the PTC, as a function of PF846 concentration (data represent mean ± s.d., n = 3 independent experiments). c, MA plot of sequences enriched for PF846-induced inhibition of translation termination, compared to translation reactions in the absence of PF846. Sequences were enriched from the mRNA library of CDH1-derived nascent chains with four amino acid positions randomized near the predicted stall site in the PTC, with only sequences containing stop codons shown. Log2-fold enrichment of sequences is plotted against total read count, for experiments carried out in duplicate. Green, sequences enriched with an adjusted P-value < 0.01. Inset, consensus sequence logo for enriched sequences 38. d, Western blot of affinity-purified stalled CDH1-derived nascent chains. Reactions in the absence or presence of 50 μM PF846 are shown. Western blot of ribosomal protein RPS19 for each sample is also shown, as a loading control. Original gels are shown in Supplementary Data Set 1. e, In vitro translation assays of the WT CDH1 stalling sequence, or with the ATHF or SFRD motif near the PTC, in the context of the CDH1 sequence (data represent mean ± s.d., n = 3 independent experiments). f, In vitro translation assays with the PCSK9 nascent chain sequence containing the HYHS motif in different positions in the absence (white bar) or presence (grey bar) of PF846. The stalled PCSK9 sequence is shown in full at the top, and the positions of the HYHS motif are indicated. Luciferase, luciferase-only control sequence. (Data represent mean ± s.d., n = 3 independent experiments). The statistical significance in panels a and c were tested by DESeq2 using the Wald test (two-sided) and adjusted for multiple comparisons by the Benjamini-Hochberg method 63. Source data for panels b, e and f are available in Supplementary Data Set 3.