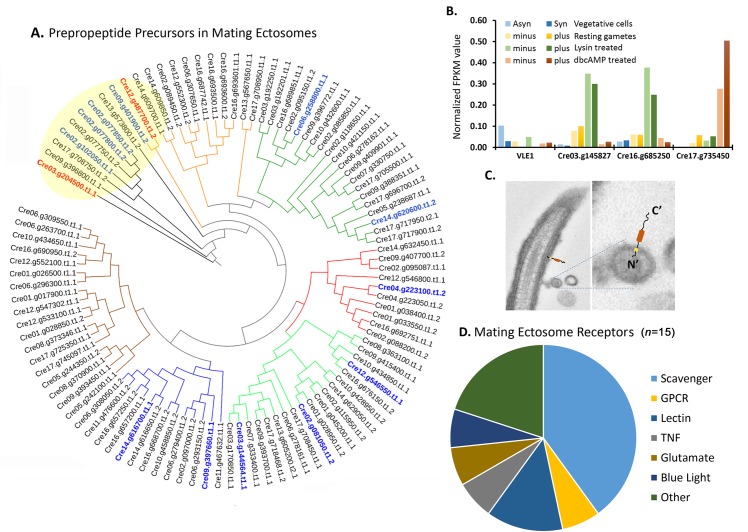
Fig 4. Proteomic analysis of mating ectosome-rich pellets reveals the presence of peptidergic signaling machinery.
A. Predicted prepropeptides (S7 Table) identified in mating ectosomes were aligned using CLUSTALW. A rooted phylogenetic tree was generated using UPGMA hierarchical clustering; groupings are delineated by colored nodes. The Cre03.g204500 chemotactic peptide precursor and the most abundant amidated product precursor (Cre12.g487700) are indicated in red; the twelve most abundant precursors are shown in blue. B. Transcriptomic analysis of the four subtilisin-like Type II membrane endoproteases identified in mating ectosomes (data from [30]). The underlying numeric data for this figure can be found in S1 Data. C. Electron micrograph of an ectosome budding from a C. reinhardtii cilium. The orientation expected of a Type II membrane protein is illustrated. D. Fifteen of the 146 C. reinhardtii receptors (S1 Table) were identified in mating ectosomes. Asyn, asynchronous; db, dibutyryl; FPKM, fragments per kilobase of transcript per million mapped reads; GPCR, G protein–coupled receptor; Syn, synchronous; TNF, tumor necrosis factor; UPGMA, unweighted paired group method with arithmetic mean; VLE1, vegetative lytic enzyme 1.