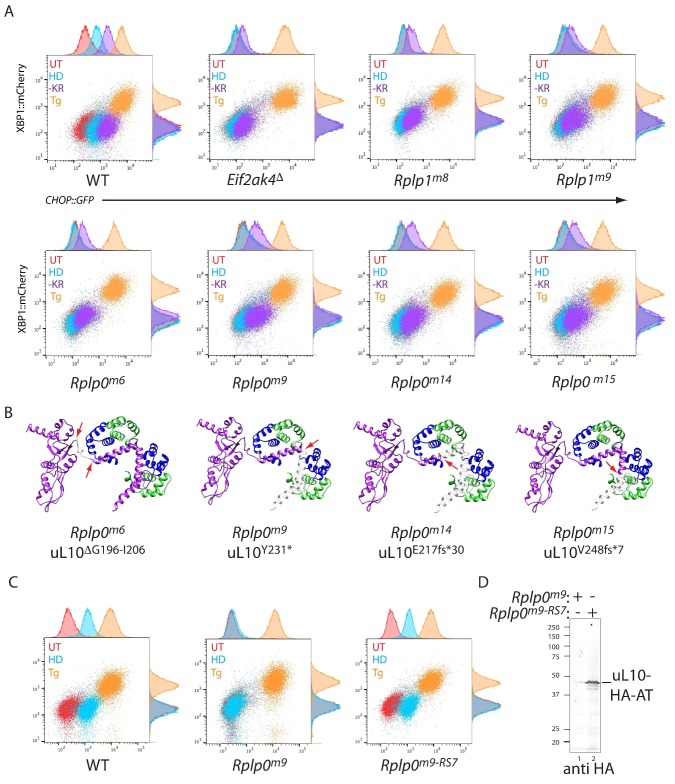
Figure 2. CRISPR-Cas9-based P-stalk lesions impair CHOP::GFP induction upon histidinol treatment or amino acid starvation.
(A) Flow cytometry analysis of the ISR-inducible CHOP::GFP and the UPR inducible XBP1::mCherry reporters from untreated (UT red), histidinol-treated (0.5 mM, HD, blue), thapsigargin-treated (1 mM, Tg, orange), or cells starved for lysine and arginine (-KR, purple) all for 20 hr. Cells were targeted with guides to the indicated genes. (B) Depiction of the genetic lesion in the Rplp0-mutant clones whose flow cytometry profile is shown above. Shown is a ribbon diagram of the P-stalk (as in Figure 1E) Red arrows indicate the boundaries of the mutations and deleted regions are colored light grey. (C) Flow cytometry analysis of parental (wildtype, WT) CHO cells, the Rplp0m9 mutant and the mutant following restoration of the P-stalk by re-targeting the mutant Rplp0 with a repair template encoding a wildtype allele with an HA epitope tag (Rplp0m9-RS7) (D) Anti-HA immunoblot of cytoplasmic extracts from the parental Rplp0m9 and rescued Rplp0m9-RS7 cells.