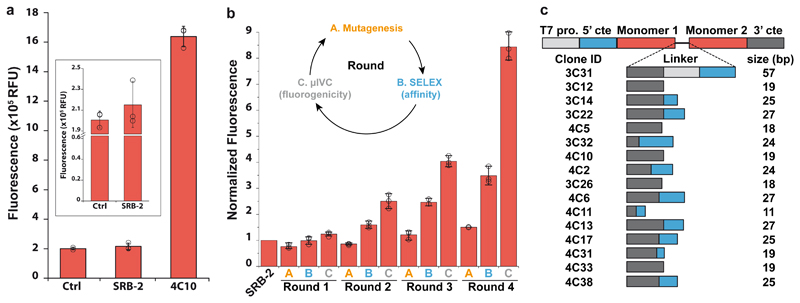
Figure 2. Isolation of Gemini-561 lighting-up aptamers by in vitro evolution.
(a) Gemini-561 activation capacity of the parental SRB-2 and the evolved 4C10 variant. 500 nM of RNAs were incubated with 50 nM of Gemini-561 and the fluorescence was measured at λ ex/em = 560/600 nm. The values are the mean of n = 3 independent experiments and the error bars correspond to ± 1 standard deviation. (b) Monitoring of the evolution process. For each round, the enriched library was transcribed in vitro in the presence of 100 nM of Gemini-561 and the fluorescence monitored. The fluorescence apparition rate was computed for each library and normalized to that of the parental SRB-2 aptamer. The inset schematizes the different steps (A, B and C) of an evolution round. The values are the mean of n = 3 independent experiments, each measurement being shown as an open circle. The error bars correspond to ± 1 standard deviation. (c) Schematic representation of genes coding for the 16 dimerized variants found among the 19 best aptamers at the end of the evolution process. For each variant, the width and the color of the box respectively inform on linker length (numerical value given on the right) and the nature of the sequence (light gray: T7 promoter, blue: 5'constant, dark grey: 3' constant). Red boxes correspond to SRB-2-derived core. The clone ID refers to the round of selection from which the clone was extracted (first number) and the clone number assigned during the final screening.