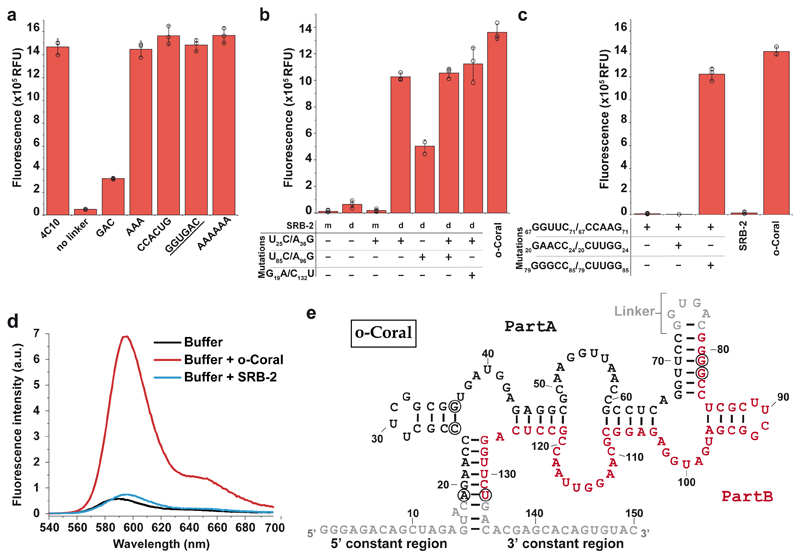
Figure 3. Characterization and engineering of the evolved molecule.
(a) Impact of linker size and sequence on the capacity of 4C10 aptamer to activate Gemini-561 fluorescence. 500 nM of RNAs were incubated with 50 nM of Gemini-561 and the fluorescence was measured at λ ex/em = 560/600 nm. The underlined sequence corresponds to o-Coral linker. (b) Contribution of the dimerization and the mutations to o-Coral functionality. SRB-2 aptamer was used as scaffold either in its monomeric (m) or dimeric (d) form containing o-Coral linker. Indicated mutations were then implemented and the different constructs tested as above. (c) Identification of interacting regions. A destabilized mutant (67GGUUC71/67CCAAG71) of o-Coral and two potential compensatory mutants (1: 67GGUUC71/67CCAAG71_20GAACC24/20CUUGG24 and 2: 67GGUUC71/67CCAAG71_79GGGCC85/79CUUGG85) were prepared and tested as above. The values (a-c) are the mean of n = 3 independent experiments, each measurement being shown as an open circle. The error bars correspond to ± 1 standard deviation. (d) Fluorescence emission spectra of Gemini-561 (200 nM) in absence and in the presence of RNA aptamers (600 nM). Excitation wavelength was 530 nm. Results were found similar in n = 3 independent experiments. (e) Model of secondary structure for o-Coral aptamer. This model was established based on enzymatic probing experiments (Supplementary Fig. 8) and mutagenesis experiments shown on c. SRB-2 derived sequences (Part A and B) are shown in black or red whereas the constant regions and the linker are shown in grey. Acquired mutations found to contribute to o-Coral function are circled in black.