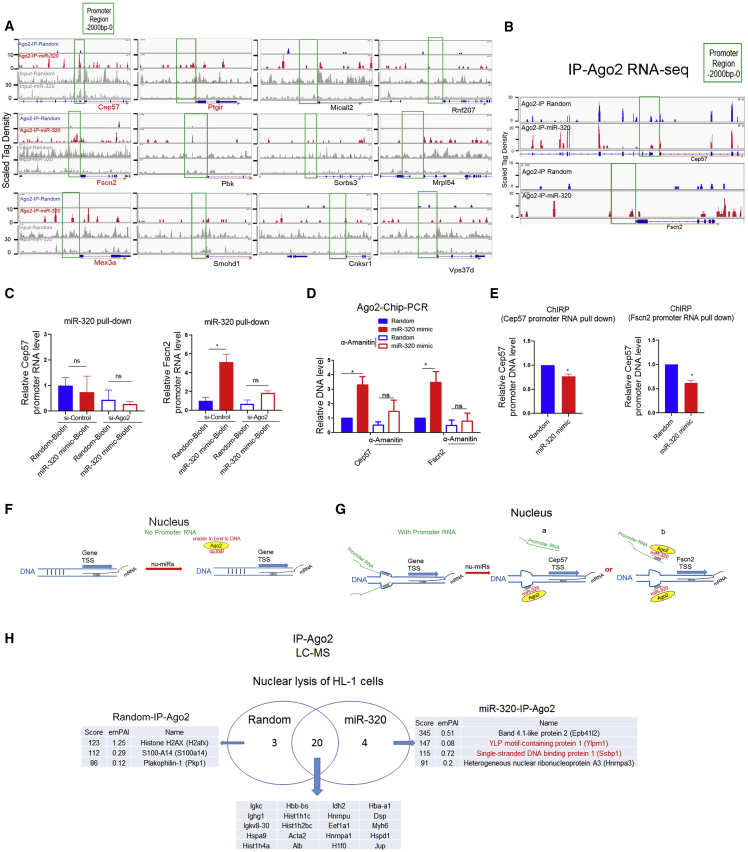
Figure 4.
Promoter ncRNAs Were Involved in miR-320-DNA Binding
(A) Ago2-ChIP-seq profiles of input DNA and immunoprecipitations in murine HL-1 cardiomyocytes transfected with miR-320 mimic or random. ChIP-seq scales were in reads per million unique mapped reads. Ago2 binding events were increased in promoter regions of Cep57, Fscn2, Mex3a, and Ptgir by miR-320. (B) Ago2-immunoprecipitation (IP) RNA-seq analysis of immunoprecipitations in H9c2 cardiomyocytes transfected with miR-320 mimic or random. RIP-seq scales were in reads per million unique mapped reads. miR-320 increased Ago2 binding on Fscn2 promoter RNA but not Cep57 promoter RNA. (C) RNA pulldown assays using streptavidin-labeled magnetic beads in HL-1 cardiomyocytes treated with the biotinylated miR-320 mimic. miR-320 pulldown showed direct targeting of miR-320 on Fscn2 promoter RNA; n = 3, *p < 0.05 by t test. (D) α-Amanitin treatment decreased miR-320-mediated Ago2 binding on gene promoters detected by Ago2-ChIP-PCR. (E) ChIRP analysis using biotinylated antisense probes to capture promoter RNAs and associated chromatin in HL-1 cells. miR-320 treatment led to detachment of Cep57 and Fscn2 promoter RNAs from their promoter DNA; n = 3, *p < 0.05 versus random by t test. (F and G) Promoter RNAs might be required for miR-320 binding on DNA. Schematic diagrams of Ago2/miRNA complexes binding to DNA without (F) or with (G) promoter RNA. Promoter RNA transcripts act as “pioneers” to disrupt chromatin that permits Ago2/miRNA complexes to target specific single-stranded DNA. (H) LC-MS analysis of Ago2-associated proteins in the nucleus.