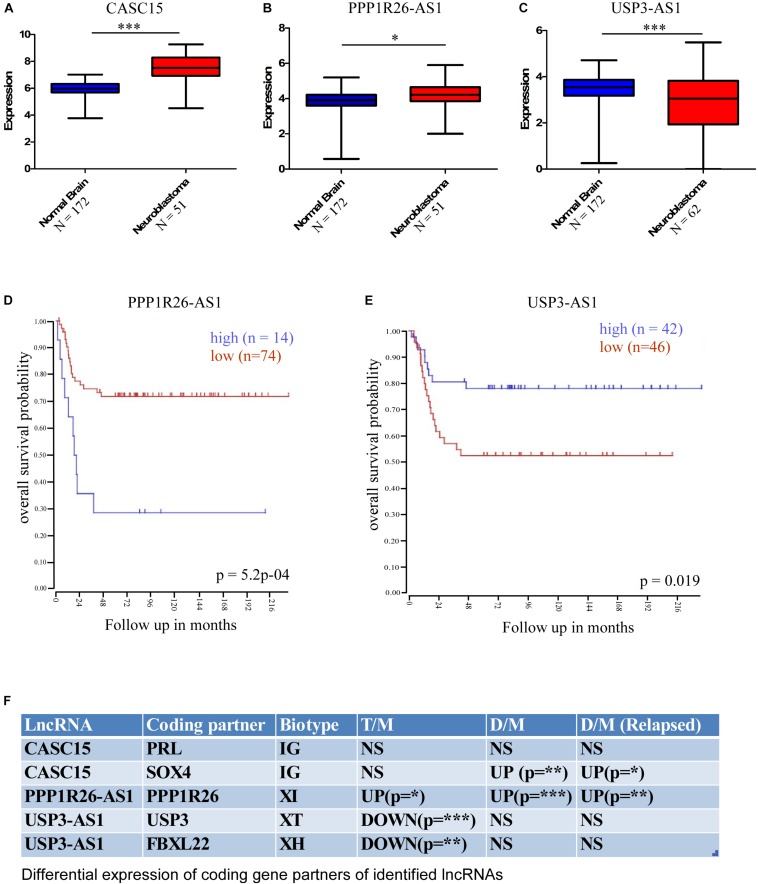
FIGURE 4.
Cross-validation of differentially expressed lncRNA expression in the cancer database and differential expression of lncRNA-associated protein-coding partners. This database revealed that the expressions of three lncRNAs were significantly altered in neuroblastoma samples; two were upregulated, (A) CASC15 (Normal brain, N = 172 and neuroblastoma samples [Hiyama], N = 51), (B) PPP1R26-AS1 (Normal brain, N = 172 and neuroblastoma samples [Hiyama], N = 51) and one was downregulated, (C) USP3-AS (Normal brain, N = 172 and neuroblastoma samples [Delattre], N = 64) (∗p ≤ 0.05, ∗∗∗p ≤ 0.0005, unpaired two-tailed t-test with Welch’s correction). Kaplan–Meier survival plots of (D) upregulated gene PPP1R26-AS1 (p = 5.2∗10– 4, number of samples with high expression = 14, number of samples with low expression = 74) and (E) downregulated gene USP3-AS1 (p = 0.019, number of samples with high expression = 42, number of samples with low expression = 46). Data were obtained from http://hgserver1.amc.nl/cgi-bin/r2/main.cgi. (F) Table showing differential expression of coding-gene partners of identified lncRNAs. LncRNA CASC15 has two coding partners, PRL and SOX4, and PRL was not significantly altered, while SOX4 was significantly upregulated in DTCs and relapsed DTCs. PPPIR26-AS1-associated coding gene PPP1R26 was significantly upregulated in all three types of neuroblastoma samples. USP3-AS1-associated coding genes USP3 and FBXL22 were significantly downregulated only in primary tumor samples (∗p ≤ 0.05, ∗∗p ≤ 0.005, ∗∗∗p ≤ 0.0005 unpaired two-tailed t-test with Welch’s correction). D/M represents DELs between DTCs and MNCs, T/M represents DELs between primary tumor and MNCs, and D/M (relapsed) represents DELs between relapsed DTCs and relapsed MNCs. IG, Intergenic lncRNA, XH, Divergent or Antisense Head-to-Head lncRNAs, XT, Convergent or Antisense Tail-to-Tail lncRNAs, XI, Antisense Inside lncRNAs.