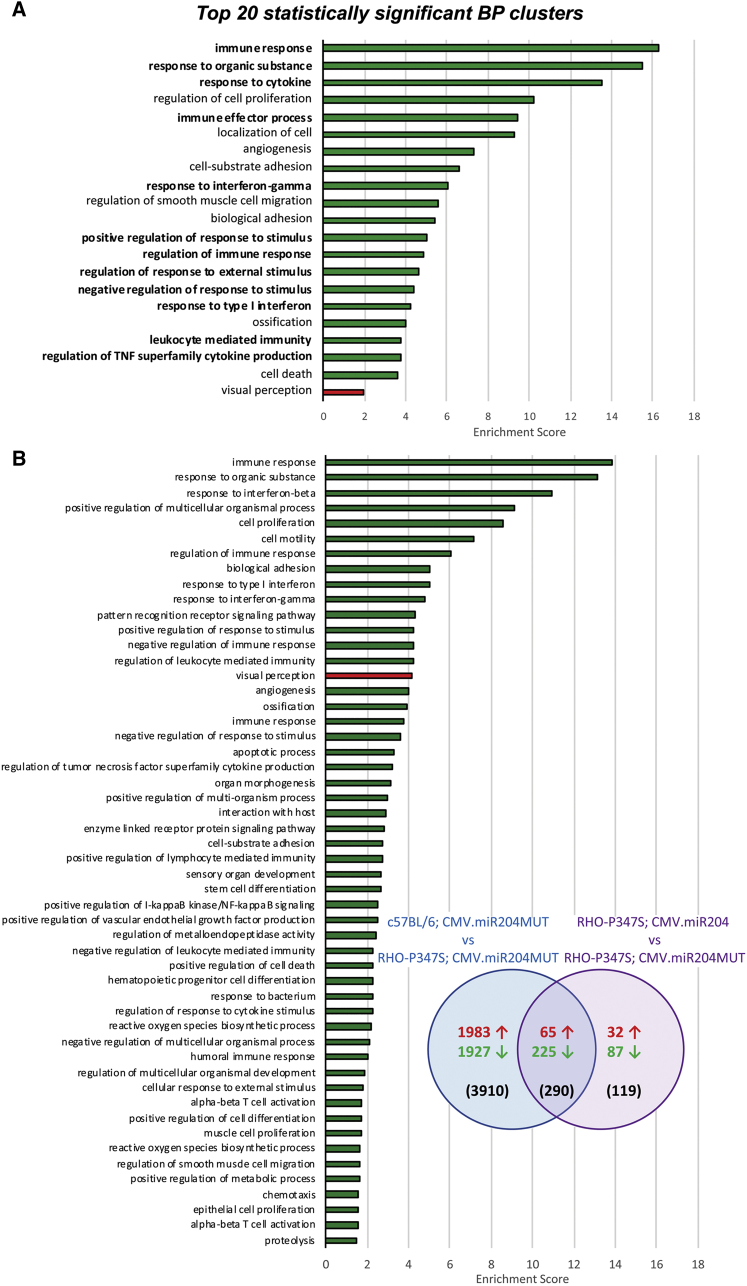
Figure 3.
Identification of the Pathways Underlying the miR-204-Protective Effect in RHO-P347S Transgenic Mice
(A) Top 20 enriched biological processes (BPs) among the differentially expressed genes in RHO-P347S retinas injected with AAV.CMV.miR204. Upregulated genes are significantly enriched for the visual perception BP (red). Downregulated genes are significantly enriched for BPs related to innate immune response, inflammatory response, defense response, and cell death, among others (green). Terms that are directly related to the ancestor terms of “immune system process” (GO: 0002376) and “response to stimulus” (GO: 0050896; ancestor of inflammatory response-related terms) are indicated in bold fonts. Enrichment score values are plotted on the x axis. (B) GOEA of the 290 genes (n = 225 downregulated + 65 upregulated) that are common between the 420 DEGs altered upon AAV-miR-204 expression in RHO-P347S (i.e., comparison of the RHO-P347S eyes injected with the CMV.miR204 versus miR204MUT control vector; purple circle) and the 4,211 DEGs between control-injected wild-type (C57BL/6) and RHO-P347S (blue circle) retinas. The 290 DEGs show the same trend of down- or upregulation in the two comparisons (11 DEGs that show opposite correlation between the two comparisons are not included). The number of DEGs that are either specific of each comparison or overlapping is shown in parentheses. In the Venn diagram, the number of upregulated genes is shown in red and that of downregulated genes in green. All significantly enriched BP terms, in which the common DEGs are involved, are shown (green bars for the downregulated DEGs; red bar for the upregulated DEGs). Terms are ranked according to the enrichment score of each cluster.