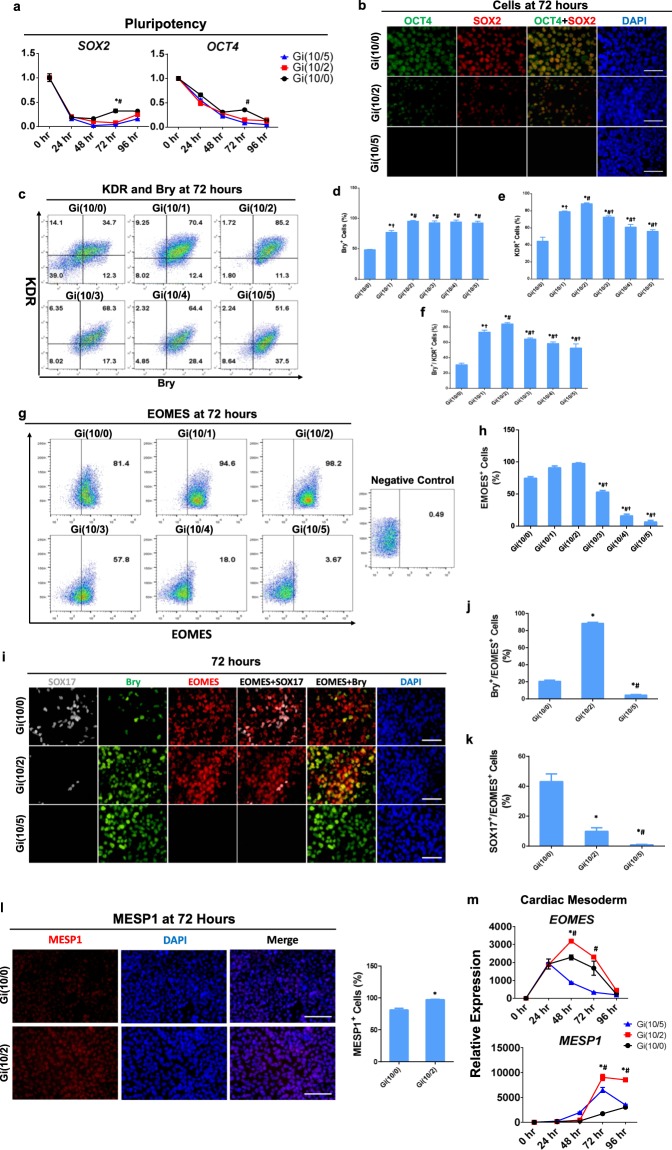
Figure 2.
The optimized Gi(I/M)Wi protocol promotes mesendodermal/mesodermal specification and limits endodermal commitment in differentiating hiPSCs. (a) mRNA levels of the pluripotency genes SOX2 and OCT4 were quantified via qRT-PCR in Gi(10/0), Gi(10/2), and Gi(10/5) cells at the indicated time points after differentiation was initiated and then normalized to GAPDH mRNA levels and to measurements taken at 0 hr (n = 3 different batches of differentiated cells). *P < 0.05 vs Gi(10/2), #P < 0.05 vs Gi(10/5). (b) Gi(10/0), Gi(10/2), and Gi(10/5) cells were collected at Hour 72 of differentiation and immunofluorescently labelled for OCT4 and SOX2 expression; nuclei were counterstained with DAPI (bars = 50 μm). (c) Expression of the mesendodermal marker Bry and the mesodermal marker KDR was evaluated in cells from all six treatment groups via flow cytometry at Hour 72 of differentiation; then, the proportion of cells that expressed (d) Bry, (e) KDR, and (f) both Bry and KDR was calculated and presented as a percentage of the total number of cells. *P < 0.05 vs Gi(10/0), #P < 0.05 vs Gi(10/1), †P < 0.05 vs Gi(10/2). (g) Expression of the mesendodermal marker EOMES was evaluated in cells from all six treatment groups via flow cytometry at Hour 72 of differentiation; then, (h) the proportion of cells that expressed EOMES was calculated and presented as a percentage of the total number of cells. *P < 0.05 vs Gi(10/0), #P < 0.05 vs Gi(10/1), †P < 0.05 vs Gi(10/2). (i) Gi(10/0), Gi(10/2), and Gi(10/5) cells were collected at Hour 72 of differentiation in a 96-well plate (3 wells per group), immunofluorescently labelled for Bry and EOMES expression and for expression of the definitive endoderm marker SOX17, and nuclei were counterstained with DAPI (bars = 50 μm); then, the proportion of cells that expressed (j) both Bry and EOMES, and (k) both SOX17 and EOMES was quantified and presented as a percentage of the total number of cells. *P < 0.05 vs. Gi(10/0). #P < 0.05 vs Gi(10/2). All experiments were repeated three times; 27–30 randomly selected fields (3–4 fields per well) from each group were evaluated. (l) Gi(10/0) and Gi(10/2) cells were collected at Hour 72 of differentiation and immunofluorescently labelled for expression of the mesodermal marker MESP1; nuclei were counterstained with DAPI (bars = 100 μm). The right bar plot showed percentage of MESP1+ cells. *P < 0.05; all experiments were repeated three times; 27–30 randomly selected fields (3–4 fields per well) from each group were evaluated. (m) mRNA levels of the definitive cardiac mesoderm markers EOMES and MESP1 were quantified via qRT-PCR in Gi(10/0), Gi(10/2), and Gi(10/5) cells at the indicated time points after differentiation was initiated and then normalized to GAPDH mRNA levels and to measurements taken at 0 hr. n = 3 different batches of differentiated cells, *P < 0.05 vs Gi(10/2), #P < 0.05 vs Gi(10/5).