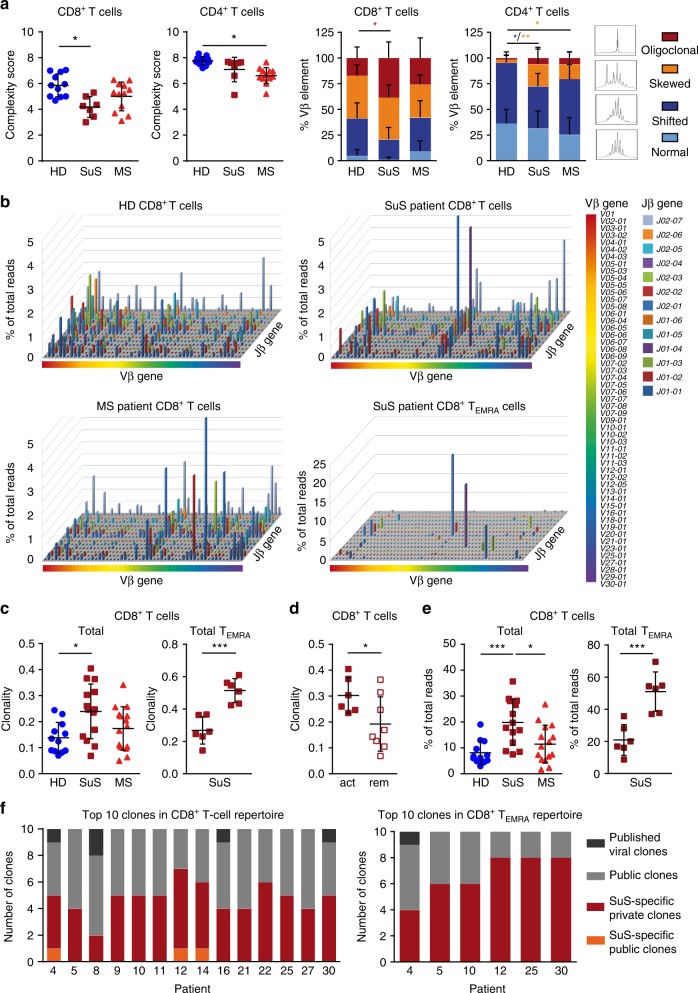
Fig. 2. Perturbations in the CD8+ TEMRA repertoire of SuS patients.
a Complexity score (left) and CDR3 length distribution (right) of peripheral CD8+ and CD4+ T cell repertoires of HD (closed blue circles, n = 11), SuS (closed cayenne squares, n = 7), and MS patients (closed red triangles up, n = 12) assessed by TCR Vβ-CDR3 spectratyping. b Representative Manhattan plots showing TCR repertoires of CD8+ T cells from a HD (top left), a SuS patient (top right, patient #10), a MS patient (bottom left), and SuS CD8+ TEMRA cells (bottom right, patient #10). The x-axis depicts all the Vβ genes, the z-axis the Jβ genes, and the column heights represent the percentage reads for each V/J gene combination. c Clonality in the CD8+ T cell repertoires of HD (closed blue circles, n = 12), SuS (closed cayenne squares, n = 14), and MS (closed red triangles up, n = 15) patients (left). Clonality in the total CD8+ T cell and CD8+ TEMRA repertoire of SuS (n = 6) patients (right). d Clonality of SuS patients with clinically active disease (closed cayenne squares, n = 6) or in clinical remission (open cayenne squares, n = 8). e Quantification of the ten most prevalent clones in the CD8+ T cell repertoires of HD (closed blue circles, n = 12), SuS (closed cayenne squares, n = 14), and MS (closed red triangles up, n = 15) patients (left), as well as in the total CD8+ T cell and CD8+ TEMRA repertoire of SuS (closed cayenne squares, n = 6) patients (right). f Graph representing the proportion of viral, public, SuS-specific public, and SuS-specific private clones in the ten most prevalent clones in the total CD8+ T cell (left) and TEMRA (right) repertoire of SuS patients. Statistical analysis was performed using Kruskal–Wallis test with Dunn’s post-test (a, c, e left graph) or unpaired Student’s t test (c right graph; d, e right graph), respectively. Error bars indicate the mean ± s.d.; p values: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Source data are provided as a Source Data file.