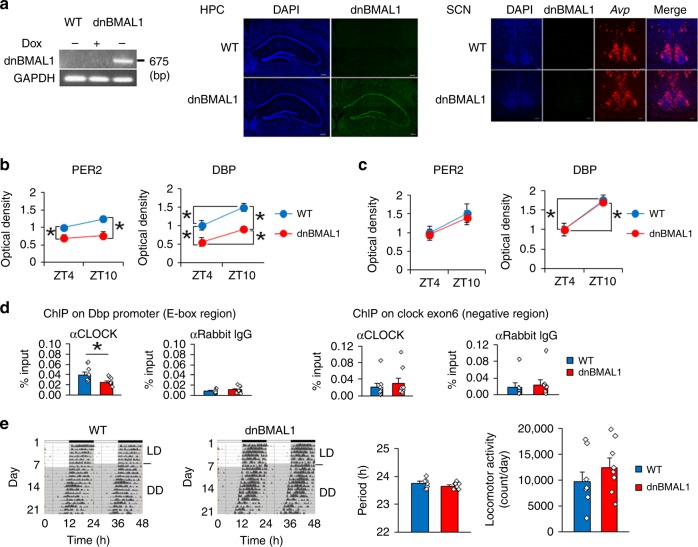
Fig. 2. Forebrain-specific inhibition of BMAL1 function.
a dnBMAL1 mRNA expression in dnBMAL1 mice. Dox-dependent dnBMAL1 mRNA expression in hippocampus (RT-PCR, left). dnBMAL1 mRNA expression in hippocampus (HPC, middle) but not SCN (right) (in situ hybridization). AVP mRNA expression as a marker of SCN. DAPI (nuclear stain, blue), dnBMAL1 (green), AVP (red). Scale bar, 200 μm (HPC) and 100 μm (SCN). b, c PER2 and expression levels (BMAL1 target genes) are reduced in hippocampal CA1 (b) but not in SCN (c), in dnBMAL1 mice at both ZT4 and 10. The graph represents fold changes compared to the expression levels in WT at ZT4. d dnBMAL1 blocks the CLOCK binding to Dbp promoter in the hippocampus of dnBMAL1 mice at ZT10. Anti-CLOCK antibody, but not anti-IgG, precipitated DBP promoter although DNA regions not containing E-box (clock gene exon 6) are comparably precipitated by anti-CLOCK antibody and anti-IgG. e Normal circadian locomotor rhythm in dnBMAL1 mice. Mice were housed in a 12 h light:12 h dark (LD) cycle then in constant darkness (DD). (Left) Representative activity records are double-plotted with each horizontal line representing 48 h. Circadian period (Middle) and daily locomotor activity (Right) under DD. All values are mean ± SEM. Individual data points are displayed as dots. *p < 0.05 as determined by two-way (b, c) or one-way (d, e) ANOVA with post hoc test. The results of the statistical analyses are presented in Supplementary Table 2. Source data are provided as a source data file.