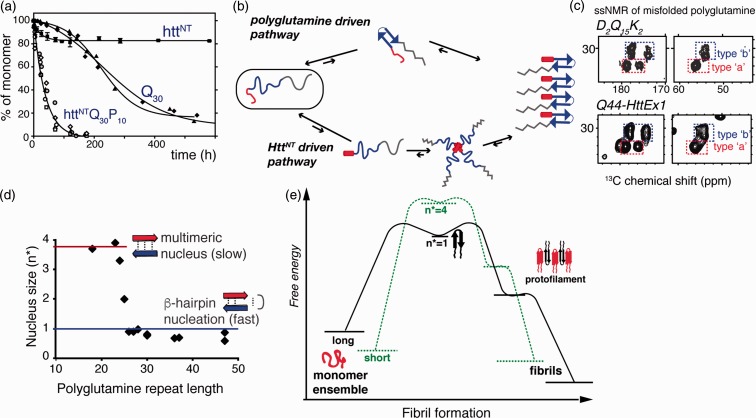
Figure 3.
Mechanisms of aggregation. (a) Expanded polyglutamine without (Q30) and with (httNTQ30P10K2) Htt flanking regions attached show dramatically different aggregation kinetics. Adapted from Sivanandam et al.77 with permission from the American Chemical Society. (b) Intrinsically disordered HttEx1 can aggregate via both polyglutamine driven (top) and HttNT- driven mechanisms (bottom). (c) Both mechanisms yield products with the same ssNMR signals for the polyglutamine fibril core structure (see Figure 4): spectra of Q44-HttEx1 and polyglutamine peptide, D2Q15K2. Adapted from Hoop et al.56. (d) Length-dependent nucleus size data96–98 show that fast aggregation of long polyglutamine is facilitated by monomeric nucleation, likely involving β-hairpin formation. (e) Schematic free energy profile of fibril formation and growth, for short and long polyglutamine, based on published nucleation and fiber elongation energy values.96,97,99,100