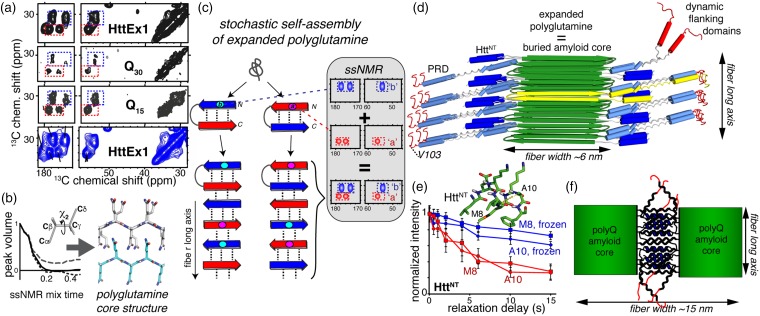
Figure 4.
SSNMR structural studies of misfolded fibrils. (a) Fibrillar polyglutamine gives an identical ssNMR signature, whether in Q44-HttEx1 fibrils (top), Q46-HttEx1 fibrils (bottom), or polyglutamine peptides with 15 or 30 glutamines. Data from Lin et al.32 and Sivanandam et al.77 Data for Q46-HttEx1 were adapted with permission from Isas et al.79 copyright 2015 American Chemical Society. (b) Tailored ssNMR dihedral angle measurements probe the polyglutamine core structure, and reveal two differently structured β-strand types (adapted from Hoop et al.56). (c) A stochastic assembly process of the alternating β-strand structures explains the ssNMR spectral signature of polyglutamine amyloid. Adapted with permission from Hoop et al.56 (d) Architecture of mutant HttEx1 fibrils derived from ssNMR and EM constraints. (e) SSNMR relaxation measurements show dynamic changes in the solvent-exposed HttNT α-helical segment upon solvent freezing. Adapted from Hoop et al.,80 with permission from the American Chemical Society. (f) Schematic illustration of flanking-domain interactions enabling higher order assembly of the wider HttEx1 fibril polymorphs shown in Figure 2(e). Panels (d) and (f) are adapted with permission from Lin et al.32 (A color version of this figure is available in the online journal.)