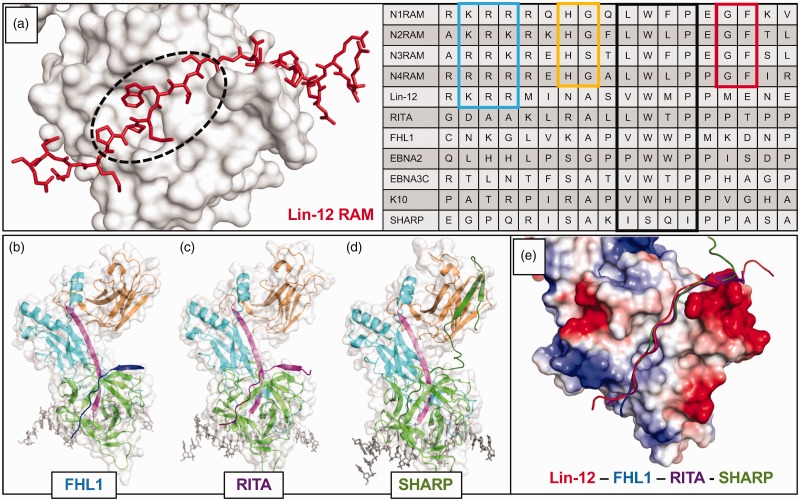
Figure 3.
β-trefoil domain (BTD) binding notch coregulators. (a) Left, Close-up view of Lin-12 RAM domain bound to the CSL BTD; PDB: 2FO1. The dashed circle shows the “hydrophobic tetrapeptide” of the RAM domain and highlights the same hydrophobic BTD region as Figure 1(c). Right, Sequence alignment of RAM and RAM-like domains of well understood BTD binding proteins. The boxed regions of sequence are: N-terminal basic patch (light blue), H[G/S] (yellow), hydrophobic tetrapeptide (black), and GF (red). Each of these is extremely well conserved in the mammalian Notch receptor RAM domains, and they can be seen to various extents in other organisms’ Notch receptors and/or other coregulators. Specifically, the hydrophobic tetrapeptide has long served as a recognition sequence for BTD binders. (b) The crystal structure of FHL1 bound to CSL, with FHL1 peptide in blue; PDB: 4J2X. (c) The crystal structure of RITA bound to CSL, with RITA in purple; PDB: 5EG6. (d) The crystal structure of SHARP bound to CSL, with SHARP in dark green; PDB: 6DKS. SHARP binds to both the BTD (green) and the CTD (orange). (e) The similarity in BTD binding architecture of different coregulators is shown in the structural alignment of the above mentioned CSL regulation complexes. The surface charge representation of CSL illustrates how the basic N-terminus of each RAM-like domain resides near a negatively charged patch of the BTD, and the hydrophobic tetrapeptide lies along the hydrophobic patch on the front face of the BTD. Despite increasingly divergent primary sequences, the peptide backbones of BTD binders align remarkably well.