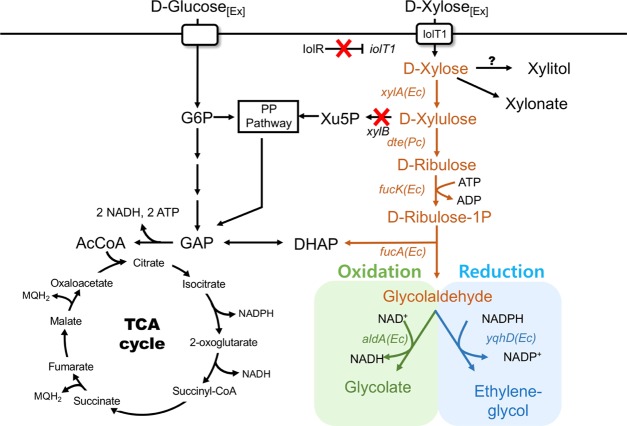
Figure 1.
Bioconversion of xylose to ethylene glycol and glycolate using engineered C. glutamicum. (A) Xylose was converted to either ethylene glycol or glycolate via synthetic metabolic pathways in C. glutamicum. The SL-1 strain (C. glutamicum ΔxylB [encoding xylulose kinase]) was used as the parental strain for the bioconversion. To convert xylose to either ethylene glycol or glycolate, five enzymatic steps (shown in colored arrows) were used by expressing genes encoding xylose isomerase (xylA) from E. coli, d-tagatose 3-epimerase (dte) from P. cichorii, l-fuculokinase (fucK) from E. coli, l-fuculose phosphate aldolase (fucA) from E. coli, aldehyde dehydrogenase (aldA) from E. coli, and aldehyde reductase (yqhD) from E. coli in SL-1. Red “X” represents gene deletion in C. glutamicum. Abbreviations: G6P, glucose 6-phosphate; DHAP, dihydroxyacetone phosphate; PP pathway, pentose phosphate pathway; Xu5P, xylulose 5-phosphate; GAP, glyceraldehyde 3-phosphate; and AcCoA, acetyl-CoA.