SUMMARY
The germline contains an immortal cell lineage that ensures the faithful transmission of genetic and, in some instances, epigenetic information from one generation to the next. Here, we show that in Caenorhabditis elegans, the small RNA 3′-2′-O-methyltransferase henn-1/HEN1 is required for sustained fertility across generations. In the absence of henn-1, animals become progressively less fertile, becoming sterile after ~30 generations at 25°C. Sterility in henn-1 mutants is accompanied by severe defects in germline proliferation and maintenance. The requirement for henn-1 in transgenerational fertility is likely due to its role in methylating and, thereby, stabilizing Piwi-interacting RNAs (piRNAs). However, despite being essential for piRNA stability in embryos, henn-1 is not required for piRNA stability in adults. Thus, we propose that methylation is important for the role of piRNAs in establishing proper gene silencing during early stages of development but is dispensable for their role in the proliferated germline.
In Brief
Svendsen et al. identify a requirement for the small RNA methyltransferase HENN-1 in germline immortality. HENN-1 is required for piRNA stability during embryogenesis but is dispensable in the adult germline, pointing to a role for piRNAs in establishing a gene regulatory network in embryos that protects the germline throughout development.
Graphical Abstract
INTRODUCTION
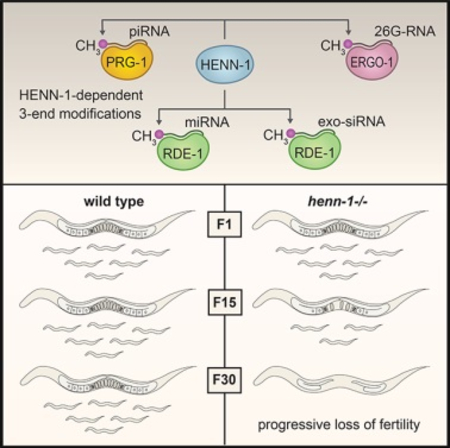
Piwi-interacting RNAs (piRNAs) have diverse roles in gene regulation and development but are best known for silencing transposons (Ozata et al., 2019). Loss of piRNAs often leads to sterility (Thomson and Lin, 2009). In Caenorhabditis elegans, however, disabling the piRNA pathway does not lead to immediate sterility but instead to a gradual loss of fertility over numerous generations (Heestand et al., 2018; Simon et al., 2014). C. elegans piRNAs initiate mRNA entry into an endogenous RNAi pathway in which small RNAs called WAGO-class 22G-RNAs are produced antisense to the mRNA target (Ashe et al., 2012; Bagijn et al., 2012; Lee et al., 2012; Luteijn et al., 2012). WAGO-class 22G-RNAs are often classified as small interfering RNAs (siRNAs), although, unlike canonical siRNAs, they are not derived from double-stranded RNA (dsRNA) and are not processed by Dicer (Ambros et al., 2003; Aoki et al., 2007; Gu et al., 2009; Pak and Fire, 2007; Sijen et al., 2007). Instead, they are produced by an RNA-dependent RNA polymerase in association with a collection of proteins called the Mutator complex (Aoki et al., 2007; Phillips et al., 2012). A subset of 22G-RNAs bind HRDE-1, a nuclear Argonaute that promotes transgenerational inheritance of RNAi and is required for transgenerational fertility (Buckley et al., 2012). The 22G-RNAs produced downstream of piRNAs, particularly those bound by HRDE-1, are thought to provide a memory of piRNA activity that seemingly persists over multiple generations, which may explain why the loss of the primary Piwi Argonaute prg-1 does not cause immediate sterility (Ashe et al., 2012; Buckley et al., 2012; Luteijn et al., 2012; Shirayama et al., 2012).
piRNAs are methylated at their 3′ ends by the 3′-2′-O-methyltransferase HEN1 (Ozata et al., 2019). First characterized in Arabidopsis, methylation protects small RNAs from 3′–5′ trimming and decay (Billi et al., 2012; Horwich et al., 2007; Kamminga et al., 2012; Kurth and Mochizuki, 2009; Li et al., 2005; Montgomery et al., 2012; Saito et al., 2007; Yu et al., 2005). Small RNAs are bound at both their 5′ and 3′ ends by the Piwi/Argonaute they associate with. The 3′ end is anchored in the PAZ domain, which likely protects the small RNA from nucleases and other base-modifying enzymes. Small RNAs with extensive target complementarity at their 3′ ends are released from the PAZ domain to facilitate base pairing with their targets (Kawamata and Tomari, 2010). In animals, piRNAs and siRNAs typically have extensive complementarity to their targets and are methylated, whereas miRNAs interact with only partial complementarity and are not methylated (Ozata et al., 2019). In Drosophila, methylation protects extensively base-paired small RNAs from target-directed trimming and tailing, thus explaining the link between target complementarity and methylation (Ameres et al., 2010). However, this simple complementarity rule for whether or not a class of small RNAs is methylated cannot be strictly applied to C. elegans, as most siRNAs, which share perfect complementarity to their targets, are not methylated (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012). The only class of small RNAs, aside from piRNAs, that is thought to be methylated in C. elegans is one of two branches of the 26G-RNA pathway involving the Argonaute ERGO-1 (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012; Ruby et al., 2006; Vasale et al., 2010).
C. elegans bearing mutations in the HEN1 ortholog henn-1 display only modest loss of fertility under normal growth conditions (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012). Here, however, we discover a critical role for henn-1 in preserving fertility and germline integrity from one generation to the next as we explore its role in piRNA, miRNA, and RNAi pathways in the worm.
RESULTS
henn-1 Is Required for Transgenerational Fertility
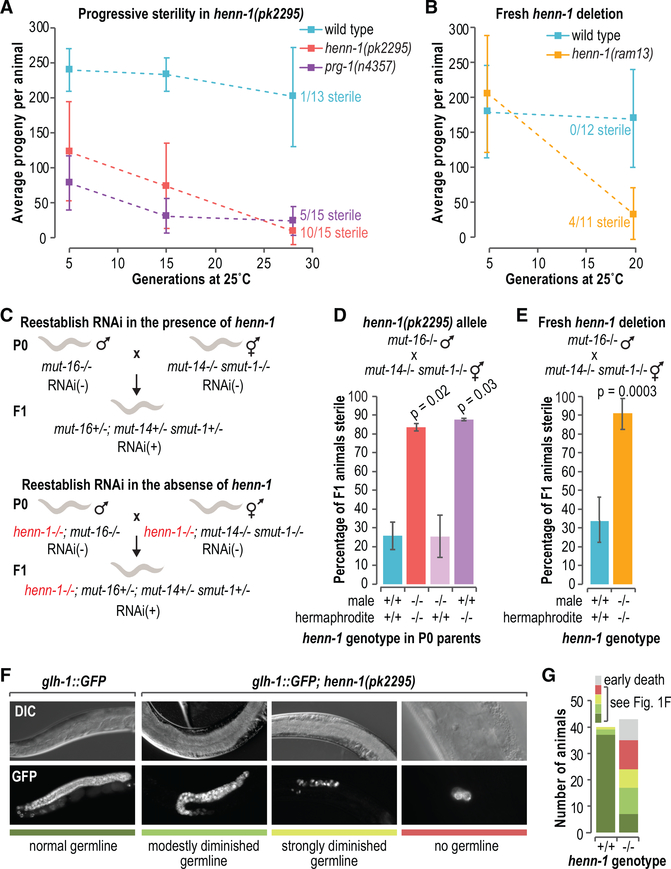
In C. elegans, the Piwi ortholog prg-1 promotes germline immortality. In the absence of prg-1, sterility ensues over ~35–80 generations (Simon et al., 2014). Given the presumed role of HENN-1 in methylating the 3′ ends of piRNAs to promote their stability, we used two separate approaches to test if, like prg-1, henn-1 is required for germline immortality. In the first set of experiments, modeled after Buckley et al. (2012), we measured the number of progeny produced from recently outcrossed (3×) henn-1(pk2295) mutants across ~28 generations at 25°C. In parallel, we measured the brood sizes of wild-type animals and freshly outcrossed (1×) prg-1(n4357) mutants as controls. At ~5 generations, the first generation at which measurements were taken, both henn-1 and prg-1 mutants displayed modest reductions in the numbers of progeny they produced relative to wild-type animals (p values = 0.00004 and 0.00002, respectively) (Figures 1A and S1A). By ~28 generations, the mean numbers of progeny produced by henn-1 and prg-1 mutants had diminished to 8 and 23, respectively, compared to 201 in wild-type (p = 0.00007 and p = 0.0002, respectively, for comparisons to wild-type), and many of the mutant animals were sterile (Figures 1A and S1A).
Figure 1. Sterility Ensues in the Absence of henn-1.
(A) Brood sizes at ~5 generations (wild-type, n = 12; henn-1, n = 17; prg-1, n = 15), ~15 generations (wild-type, n = 10; henn-1, n = 15; prg-1, n = 13), and ~28 generations (wild-type, n = 13; henn-1, n = 15; prg-1, n = 15). The numbers of sterile animals at ~28 generations are shown. p < 0.002 for all comparisons to wild-type (see Figure S1A). Error bars report standard deviation.
(B) Brood sizes at ~5 generations (wild-type, n = 14; henn-1, n = 12) and ~20 generations (wild-type, n = 12; henn-1, n = 11). The numbers of sterile animals at ~20 generations are shown. Individual data points and p values are in Figure S1B. Error bars report standard deviation.
(C) The genetics-based approach used to reestablish WAGO-class 22G-RNA production and, thus, endogenous RNAi in henn-1+/+ or henn-1−/− animals. RNAi competency is indicated by RNAi+ or RNAi–.
(D) Mean percentage of F1 progeny from the crosses illustrated in (C) that were sterile (n = 2 biological replicates each assaying the progeny from 7–27 crosses). Error bars report standard deviation.
(E) Same as in (D) but with the henn-1(ram13) allele. The henn-1 genotype is indicated and because both parents are homozygous mutant or wild-type for henn-1, the genotype applies to both the P0 parents and the F1 progeny (n = 4 independent experiments each assaying the progeny from 11–34 crosses). Error bars report standard deviation.
(F) Differential interference contrast (DIC) and GFP fluorescence images of the diminutive germlines of sterile glh-1::GFP; henn-1(pk2295) animals. An image of the germline of a glh-1::GFP animal wild-type for henn-1 is shown as a control. Strains were grown at 25°C for 10 generations. The colored bars under the images relate to the key in (G).
(G) Numbers of henn-1+/+ and henn-1−/− animals displaying germline defects exemplified in (F), as indicated in the key.
See also Figure S1.
The NL4415[henn-1(pk2295)] strain used in these experiments has been propagated over numerous generations and may have acquired background mutations that could contribute to defects in fertility. Thus, we repeated the brood size experiments with a fresh henn-1 deletion allele generated using CRISPR-Cas9. The new henn-1(ram13) allele displayed fertility rates indistinguishable from wild-type animals at ~5 generations when grown at 25°C (p = 1.0). After ~20 generations, however, animals containing the fresh henn-1 deletion produced only 33 progeny/animal on average and exhibited a high incidence of sterility, whereas wild-type animals produced 169 progeny on average and were all fertile (p = 0.0009 for comparison to wild-type) (Figures 1B and S1B).
We then used a second assay for transgenerational fertility, modeled after Simon et al. (2014), in which we followed the fertility of 10 independent henn-1(pk2295) and prg-1(n4357) mutant lines, scoring at every generation whether or not each line was fertile. All of the henn-1 mutant lines had become sterile by 29 generations at 25°C (Figure S1C). However, sterility occurred after only 12 generations in prg-1 mutants (Figure S1C). Additionally, prg-1 mutants displayed progressive sterility at 20°C across 30 generations, whereas henn-1 mutants did not (Figure S1D).
To determine if the 3′-2′-O-methyltransferase activity of henn-1 is required for transgenerational fertility, we tested whether animals containing a mutation that disrupts the catalytic site of henn-1 also display a progressive decline in fertility. Indeed, between 1 and 10 generations at 25°C, henn-1(pk2452) mutants displayed an ~55% reduction in the mean number of progeny they produced (p = 0.0072) (Figure S1E). Finally, to determine if defects in piRNA function are likely responsible for the transgenerational sterility of henn-1 mutants, we assessed whether the loss of the other class of small RNAs presumed to be methylated in C. elegans, ERGO-1-class 26G-RNAs, affects fertility. Mutations in ergo-1 did not cause transgenerational sterility (Figures S1C and S1D). Therefore, our results suggest that henn-1, and hence presumably methylation, is important for the role of piRNAs in maintaining germline immortality, although it is possible that henn-1 has additional unknown roles that contribute to its phenotype.
Sterility Ensues after Reestablishing WAGO-Class 22G-RNA Production in the Absence of henn-1
Disabling both the piRNA pathway and the WAGO-class 22G-RNA pathway and then restoring just the WAGO pathway causes sterility, despite neither pathway being essential for fertility under normal conditions (de Albuquerque et al., 2015; Phillips et al., 2015). In the animals in which the WAGO pathway is restored in the absence of piRNAs, essential genes are misrouted into the RNAi pathway for silencing (de Albuquerque et al., 2015; Phillips et al., 2015). It is not clear if this role for piRNAs in preventing silencing of essential genes is related to its role in maintaining germline immortality. However, given that henn-1 is required for piRNA stability, we tested if it is also required for fertility upon reestablishing the WAGO pathway. To reestablish the WAGO pathway, we crossed two strains containing distinct mutations, mut-16 or mut-14 smut-1, that cause loss of WAGO-class 22G-RNAs, such that their progeny received functional copies of mut-14 and smut-1 from one parent and a functional copy of mut-16 from the other parent. Thus, although neither parent was capable of WAGO-class 22G-RNA formation, the pathway was restored in their progeny (Figure 1C). If henn-1 is required for the role of piRNAs in establishing proper RNA silencing, we would predict that introducing a henn-1 mutation into the two strains used to reestablish the WAGO pathway would lead to sterility in the offspring of the cross, as we previously showed occurs when the two strains contain mutations in prg-1 (Phillips et al., 2015). Indeed, the vast majority of henn-1 mutants in which WAGO-class 22G-RNA production was restored were sterile, whereas, in the presence of henn-1, most animals were fertile (p = 0.02 for the differences between henn-1+/+ and henn-1−/−) (Figure 1D).
Reactivating WAGO-class 22G-RNA production causes sterility specifically in the absence of maternally contributed prg-1 (de Albuquerque et al., 2015; Phillips et al., 2015). To determine if, similarly, a maternal contribution of henn-1 is specifically required for fertility when reestablishing WAGO-class 22G-RNA production, we set up crosses to reactivate the WAGO pathway in which only one parent was mutant for henn-1. The progeny of animals that received a paternal, but not a maternal, contribution of wild-type henn-1 were mostly sterile (Figure 1D). In contrast, progeny that received a maternal contribution of wild-type henn-1, regardless of whether they received a paternal contribution of wild-type henn-1, were mostly fertile, indicating that, like prg-1, a maternal contribution of henn-1 is required for fertility when reestablishing WAGO-class 22G-RNA production (Figure 1D). It is unlikely that background mutations are responsible for the sterility that ensues after restoring the WAGO pathway, as both old and new alleles of henn-1 displayed similar proportions of sterile animals (~83%–90%) (Figures 1D and 1E).
We next assessed whether henn-1 mutants display germline atrophy or defects in germ cell proliferation similar to those exhibited by prg-1/piwi mutants as they go sterile either over multiple generations or upon reestablishing WAGO-class 22G-RNA production (de Albuquerque et al., 2015; Heestand et al., 2018; Phillips et al., 2015). To do so, we introduced a germ cell marker, glh-1::GFP (Andralojc et al., 2017), into henn-1(pk2295) mutants and examined adult animals after several generations at 25°C. At ten generations, many of the glh-1::GFP; henn-1(pk2295) mutants were sterile and displayed diminutive and underproliferated germlines reminiscent of sterile prg-1 mutants (Figures 1F and 1G) (de Albuquerque et al., 2015; Heestand et al., 2018; Phillips et al., 2015). Thus, henn-1 is required for proper maintenance of the germline over multiple generations. That henn-1 and prg-1 have a similar impact on germline integrity is further evidence that henn-1 is important for the role of piRNAs in preserving generational fertility.
henn-1 Impacts WAGO-Class 22G-RNA Formation throughout Development
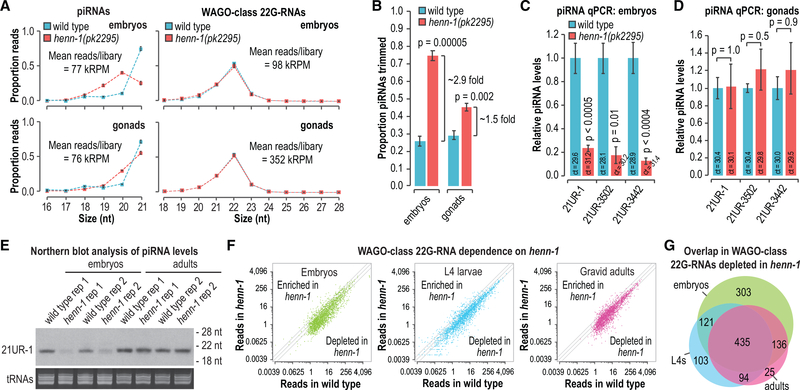
henn-1 is required for piRNA accumulation in embryos and L1 larvae but mostly dispensable later in development (Billi et al., 2012). This suggests that methylation may not be required for piRNA stability in the proliferating germline. To assess the impact of henn-1 on piRNA stability in the germline, we sequenced small RNAs from dissected distal gonads of adult animals, which are comprised primarily of germ cells (Pazdernik and Schedl, 2013), from wild-type and henn-1 mutants. We also sequenced small RNAs from mixed-stage embryos, which have a variable number of somatic cells but contain only two germline precursor cells. To determine if piRNAs are more susceptible to decay in the germline precursor cells of embryos compared to the proliferated germlines of adults, an indicator of which is trimming or truncation at the 3′ ends, we calculated the proportions of piRNA reads that displayed 1- to 5-nt truncations in our high-throughput sequencing libraries. In henn-1 mutant embryos, we observed an ~2.9-fold increase in the proportion of piRNAs that were truncated relative to wild type (p = 0.00005), whereas in gonads, we observed a more modest 1.5-fold increase (p = 0.002) (Figures 2A and 2B). piRNA trimming in gonads was almost exclusively limited to 1 nt, whereas in embryos we observed 1- to 2-nt truncations (Figure 2A). To determine if the elevated levels of piRNA truncations in henn-1 mutants corresponded to a reduction in mature piRNA levels, we used TaqMan qRT-PCR, to measure the levels of three piRNAs. Unlike high-throughput sequencing library preparation, TaqMan small RNA qRT-PCR is not thought to be hindered by 3′ methylation. Each of the piRNAs tested was strongly depleted in henn-1 mutant embryos, indicating that the relatively high levels of truncations we observed in embryos were coincident with their decay (p ≤ 0.01) (Figure 2C). In contrast, we did not detect any differences in piRNA levels in henn-1 mutant gonads, despite the modest increase we observed in piRNA trimming (p = 0.5–1.0) (Figure 2D). To confirm our qRT-PCR results, we examined 21UR-1 by northern blot. Consistent with our qRT-PCR results, 21UR-1 levels were strongly depleted in henn-1-mutant embryos but appeared unchanged in henn-1-mutant adults relative to wild-type animals (Figure 2E). Our results suggest that henn-1, and thus presumably methylation, is important for protecting piRNAs from decay in early embryos but is largely dispensable in the proliferated germline of adult animals.
Figure 2. Requirement for henn-1 in piRNA Stability and WAGO-Class 22G-RNA Production.
(A) The proportions of total high-throughput sequencing reads for piRNAs and WAGO-class 22G-RNAs in embryos and dissected gonads are plotted by size. Mean reads per library are shown as thousand reads per million total mapped reads (kRPM) (n = 3 biological replicates). Error bars report standard deviation.
(B) The proportions of total piRNA reads that display 1- to 5-nt truncations (mean trimmed reads/total reads) in embryos and dissected gonads (n = 3 biological replicates). Error bars report standard deviation.
(C and D) qRT-PCR assay of three piRNAs in embryos (C) and dissected gonads (D) (n = 3 biological replicates for each assay). The mean cycle threshold (ct) value is shown for each assay. piRNA ct values were normalized to miR-35 ct values. Error bars report standard deviation.
(E) Northern blot of the piRNA 21UR-1 in embryos and adults. Ethidium bromide (EtBr)-stained tRNAs are shown as a loading control. Two of three biological replicates are shown.
(F) Normalized WAGO-class 22G-RNA reads (reads per million total mapped reads) in wild type (x axis) and henn-1(pk2295) (y axis). Data from embryos, L4 larvae, and gravid adults are shown (no biological replicates).
(G) Venn diagram displaying the overlap in genes depleted of WAGO-class 22G-RNAs by >50% in henn-1(pk2295) embryos, L4 larvae, and adults.
See also Figure S2.
piRNAs trigger secondary siRNA (WAGO-class 22G-RNA) production from their targets, and consequently, 22G-RNA production from many WAGO target genes is reduced in prg-1 mutants (Lee et al., 2012). To identify the requirement for henn-1 in triggering WAGO-class 22G-RNA production, we subjected small RNAs from wild-type and henn-1(pk2295) mutant embryos, L4 larvae, and gravid adults to high-throughput sequencing. In each of the developmental stages, we observed a similar modest loss of 22G-RNAs from WAGO targets in henn-1 mutants (Figure 2F). Consistent with previous studies, WAGO-class 22G-RNAs did not display 3′ truncations in the absence of henn-1, indicating that they are indirectly affected by loss of henn-1 activity (Figure 2A) (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012). A total of ~75% of WAGO targets depleted of 22G-RNAs in henn-1 mutant adults were also depleted of 22G-RNAs in prg-1 mutants, indicating that henn-1 primarily affects piRNA-dependent 22G-RNAs (Figure S2) (Phillips et al., 2015). The majority of WAGO target genes depleted of 22G-RNAs >50% in embryos were also depleted of 22G-RNAs >50% in adults and L4 stage larvae (Figure 2G). These results suggest that henn-1 impacts piRNA-dependent 22G-RNA production throughout development despite the requirement for henn-1 in piRNA stability being restricted to early stages of development.
Loss of henn-1 Has Little Impact on Gene Expression in the Germline
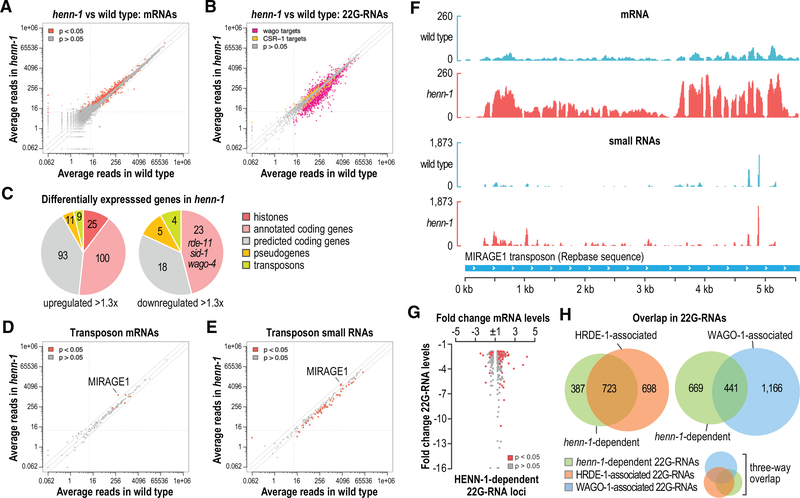
Because the loss of henn-1 has a modest impact on the health of animals grown at 20°C (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012), we could examine its role in regulating gene expression in the germline, where piRNAs are active, without developmental defects confounding the results. However, a caveat to this approach is that henn-1 is not required for piRNA stability in adults, the stage at which we are able to dissect germline tissue. Nonetheless, because many piRNA-dependent 22G-RNAs are depleted in henn-1 mutant adults at 20°C, we could the assess the impact of their loss on germline gene expression. Therefore, we did mRNA sequencing using the same wild-type and henn-1(pk2295) RNA samples from adult gonads that were used for the small RNA high-throughput sequencing analysis described above (Figures 2A and 2B). We then performed differential gene expression analysis applying an arbitrary 1.3-fold change cutoff, which excludes many genes that are differentially expressed based on a false discovery rate of 0.05 but which increases the likelihood that differences observed are biologically relevant. As we predicted based on the health of the animals at 20°C, only modest changes in mRNA expression were observed in henn-1 mutants relative to wild-type animals, with 238 genes significantly upregulated and 50 genes significantly downregulated >1.3-fold (Figure 3A; Table S1).
Figure 3. Differential Expression of mRNAs and Small RNAs in henn-1 Gonads.
(A) Scatterplot displaying each gene as the average number of normalized reads in wild type (x axis) and henn-1(pk2295) (y axis) (n = 3 biological replicates). Grey lines are at −2-, 1-, and 2-fold change relative to wild type. Dashed lines are at 10 reads on each axis.
(B) Scatterplot displaying each WAGO and CSR-1 target gene as the average number of normalized small RNA reads in wild type (x axis) and henn-1(pk2295) (y axis) (n = 3 biological replicates).
(C) Pie charts displaying the proportions of genes upregulated or downregulated in henn-1(pk2295) (p < 0.05) belonging to each of the indicated classes.
(D) Scatterplot displaying each transposon family as the average number of normalized mRNA reads in wild type (x axis) and henn-1(pk2295) (y axis) (n = 3 biological replicates).
(E) Scatterplot displaying each transposon family as the average number of normalized small RNA reads in wild type (x axis) and henn-1(pk2295) (y axis) (n = 3 biological replicates).
(F) mRNA and small RNA read distribution across the MIRAGE transposon consensus sequence. One of three biological replicates is shown.
(G) Scatterplot displaying fold change in mRNA levels in henn-1(pk2295) relative to wild type for each gene depleted of 22G-RNAs >1.3-fold in henn-1(pk2295).
(H) Venn diagrams displaying the overlap between genes depleted of 22G-RNAs in henn-1(pk2295) and genes that yield 22G-RNAs enriched >1.3-fold in WAGO-1 or HRDE-1 coIPs. The three-way Venn diagram is a combination of the two-way Venn diagrams.
In parallel, we performed differential expression analysis of small RNAs from these samples, which we had earlier used in our trimming analysis, to assess more comprehensively the impact of henn-1 on 22G-RNA levels. We identified 1,084 WAGO target genes that were depleted of 22G-RNAs >1.3-fold (Figure 3B; Table S2). A small subset of genes targeted by CSR-1, an Argonaute implicated in licensing genes for expression (Claycomb et al., 2009; Seth et al., 2013; Wedeles et al., 2013), also displayed altered levels of 22G-RNAs (Figure 3B; Table S2). piRNA levels in our sequencing data appeared elevated in henn-1 mutants, likely due to increased capture efficiency, as 3′-2′-O-methylation inhibits RNA ligation and reverse transcription during library preparation, despite our efforts to minimize such biases (Table S2) (Munafó and Robb, 2010).
Among the upregulated genes, 20% are histones, representing each of the core families—H2A, H2B, H3, and H4 (Figures 3C and S3A; Table S1). As these are multicopy gene families and the effects are modest (1.35- to 1.86-fold), it is unclear what impact this might have on the chromatin landscape. A small subset of histone genes had reduced levels of 22G-RNAs in henn-1 mutants, suggesting that henn-1-dependent 22G-RNAs may have a role in regulating histones, although it is possible that altered cell physiology in henn-1 mutants indirectly impacts histone levels (Figure S3A; Table S2). The levels of several genes involved in RNAi—sid-1, rde-11, and wago-4—were modestly reduced in henn-1 mutants, each of which also had reduced levels in henn-1 in a microarray-based study using whole animals (Figures 3C and S3B–S3D; Table S1) (Kamminga et al., 2012). sid-1 and rde-11 are required for optimal exogenous RNAi, and their downregulation could explain the reduced RNAi efficiency previously observed in henn-1 mutants (Billi et al., 2012; Kamminga et al., 2012; Winston et al., 2002; Yang et al., 2012; Zhang et al., 2012). WAGO-4 transports siRNAs from parent to progeny to promote multigenerational RNAi, and thus, its misregulation may have a role in the transgenerational sterility of henn-1 mutants (Wan et al., 2018; Xu et al., 2018). However, wago-4 levels were reduced by only ~1.3-fold in henn-1 mutants, suggesting that other factors likely contribute to the transgenerational sterility of henn-1 mutants.
As piRNAs are well known for their role in regulating transposons, we used a more targeted approach to assess the role of henn-1 in transposon regulation. We aligned our RNA sequencing (RNA-seq) reads from wild-type and henn-1 mutants to each of 152 transposon family consensus sequences and compared the levels of small RNAs and mRNAs produced from each family (Jurka, 2000). Only five transposon families were misregulated in henn-1 mutants, most notably MIRAGE, which was upregulated ~4-fold and is also upregulated in prg-1 mutants (Figure 3D; Table S3) (McMurchy et al., 2017). Desilencing of MIRAGE is partially responsible for the sterility of several heterochromatin factor mutants linked to small-RNA-mediated gene silencing (McMurchy et al., 2017). Most 22G-RNAs produced from MIRAGE were upregulated in henn-1, contrary to what would be expected if these siRNAs were involved in RNA silencing (Figures 3E and 3F; Table S3). It is possible that MIRAGE is indirectly affected by henn-1, although given that it is also upregulated in prg-1 mutants, it may be that other elements of the pathway obscure the relationship between 22G-RNA production and RNA silencing. We, therefore, examined the relationship between henn-1-dependent 22G-RNAs and mRNA misregulation in henn-1 mutants more generally. Genes depleted of 22G-RNAs in henn-1 mutants had a tendency to be upregulated, although there was considerable variation and some were instead downregulated (Figure 3G). This inconsistency could be due to complex feedback between mRNA expression and 22G-RNA production. The numbers of predicted piRNA target sites in mRNAs misregulated in henn-1 mutants, as determined by sequence complementarity and by PRG-1-mRNA co-immunoprecipitation (coIP) (CLASH), were similar between genes that were upregulated and those that were downregulated (Table S1) (Shen et al., 2018; Wu et al., 2019; Zhang et al., 2018). However, all but one misregulated gene had multiple predicted piRNA target sites in at least one of the two target prediction methods, and it is not clear which genes are bona fide piRNA targets (Table S1). Further studies exploring the relationship between piRNA-mRNA interactions and gene expression may shed light on these results.
HRDE-1 is a nuclear Argonaute that associates with a subset of WAGO-class 22G-RNAs and is required for RNAi inheritance, piRNA-mediated transgenerational gene silencing, and at 25°C for transgenerational fertility (Ashe et al., 2012; Buckley et al., 2012; Luteijn et al., 2012; Shirayama et al., 2012). Because henn-1 is also required for transgenerational fertility at 25°C and for piRNA stability and may, therefore, promote the production of HRDE-1-associated 22G-RNAs, we examined the overlap between 22G-RNAs that are henn-1-dependent and those that associate with HRDE-1 or with WAGO-1, a WAGO Argonaute not implicated in transgenerational inheritance. Of the ~1,100 genes depleted of 22G-RNAs in henn-1 mutants, ~66% were enriched for 22G-RNAs that co-immunoprecipitated (coIP’d) with a FLAG::HRDE-1 fusion protein (Figure 3H; Table S4). Only ~40% were enriched for 22G-RNAs that coIP’d with GFP::FLAG::WAGO-1, the majority of which also coIP’d with HRDE-1 (Figure 3H; Table S4).
Together, our results indicate that henn-1 is important for the production of piRNA-dependent and HRDE-1-associated 22G-RNAs. However, the consequence of the loss of henn-1 on gene expression in the adult germline is modest under the optimal growing conditions in which our experiments were done. Given that growth at elevated temperatures over multiple generations exacerbates the henn-1 phenotype, more dramatic defects in gene expression almost certainly underlie the progressive sterility of henn-1 mutants. Unfortunately, the severe germline defects that occur under such conditions would complicate germline dissections and confound RNA-seq analysis of gene expression.
Exogenous siRNAs Are Modified at Their 3′ Ends
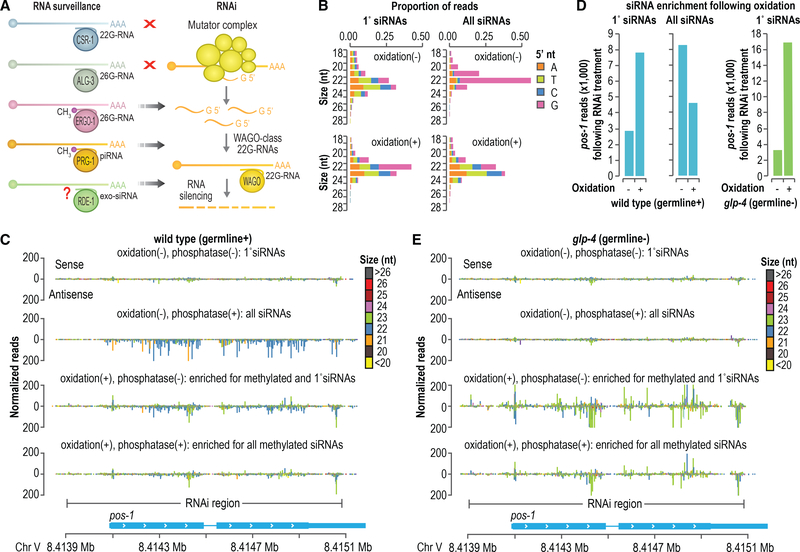
Both classes of small RNAs that are presumably methylated in C. elegans, piRNAs, and ERGO-1-class 26G-RNAs are thought to function primarily to trigger WAGO-class 22G-RNA production (Figure 4A) (Billi et al., 2014). Thus, we hypothesized that methylation is specific to small RNAs that act upstream of the WAGO pathway. Exogenous siRNAs produced from dsRNA administered during RNAi treatment bind to the Argonaute RDE-1 and like piRNAs and ERGO-1-class 26G-RNAs act as primary small RNAs to trigger 22G-RNA production (Pak and Fire, 2007; Sijen et al., 2007; Tabara et al., 2002). If methylation is specific to primary small RNAs upstream of WAGO-class 22G-RNAs, exogenous siRNAs should also be methylated. To address this, we did RNAi against the endogenous gene pos-1 in wild-type animals and tested whether primary and secondary siRNAs (i.e., WAGO-class 22G-RNAs) corresponding to pos-1 were modified at their 3′ ends by using a high-throughput sequencing approach. To enrich for small RNAs containing 3′ modifications, a fraction of the small RNA sample from animals undergoing pos-1 RNAi was treated with sodium periodate, an oxidizing agent that disrupts the 3′ base of unmethylated, but not 2′-O-methylated, small RNAs, thereby inhibiting adaptor ligation to the 3′ end (Seitz et al., 2008). Although sodium periodate treatment does not directly assess methylation, it is a widely used proxy that is not known to be confounded by other small RNA chemical modifications (Yu and Chen, 2010). The other fraction was treated similarly but was not oxidized. Each of the two fractions was further split between an RNA 5′ polyphosphatase treatment, which reduces the triphosphate group found on 22G-RNAs to monophosphate, thereby enabling capture by 5′ ligation during small RNA library preparation (Pak and Fire, 2007; Sijen et al., 2007), and a control treatment. The fractions treated with RNA 5′ polyphosphatase would presumably yield mostly 22G-RNAs, as this class of small RNAs is far more abundant than primary siRNAs (Pak and Fire, 2007; Sijen et al., 2007). The fractions that were not treated with RNA 5′ polyphosphatase would likely exclude 22G-RNAs because of an adaptor-ligation-incompatible 5′ triphosphate group. The four small RNA fractions were subjected to library preparation and high-throughput sequencing.
Figure 4. Primary siRNAs Are Modified at Their 3′ Ends.
(A) Model depicting the interplay between RNA surveillance and WAGO-class 22G-RNA production by the RNAi pathway.
(B) Size distribution and 5′ nucleotide composition of pos-1 siRNAs following pos-1 RNAi, as determined by high-throughput sequencing. Small RNAs were treated with sodium periodate (oxidation) to enrich for methylated small RNAs or subjected to a control treatment prior to library preparation. The “1° siRNAs” plot corresponds to libraries in which the small RNAs were not treated with RNA 5′ polyphosphatase and, thus, do not include di- and tri-phosphorylated small RNAs (e.g., 22G-RNAs). The “All siRNAs” plot corresponds to libraries in which the small RNAs were treated with RNA 5′ polyphosphatase and, thus, include both 1° and 2° siRNAs.
(C) Small RNA read distribution across pos-1 from wild-type high-throughput sequencing libraries in which the small RNAs were treated with sodium periodate (oxidation +) and RNA 5′ polyphosphatase (polyphosphatase +) as indicated.
(D) Bar plots displaying normalized reads (reads per million total mapped reads) mapping to pos-1 following sodium periodate (oxidation +) or control treatment (oxidation –). The plots on the left are from wild-type animals, and the plot on the right is from glp-4(bn2) mutants, which lack proliferated germlines and for which only primary siRNAs are shown.
(E) Small RNA read distribution across pos-1 from glp-4(bn2) high-throughput sequencing libraries in which the small RNAs were treated with sodium periodate (oxidation +) and RNA polyphosphatase (polyphosphatase +) as indicated.
See also Figure S4.
Small RNAs derived from pos-1 in both oxidized and non-oxidized libraries in which the small RNAs were not treated with RNA 5′ polyphosphatase displayed features associated with primary siRNAs: similar proportions of 22- and 23-nt species, little or no bias for a 5′ G, and production from both sense and antisense strands of the dsRNA administered to the animals to elicit RNAi (Figures 4B and 4C) (Zhang et al., 2012). These presumptive primary siRNAs were enriched >2-fold by oxidation, indicating that at least a subset is modified at their 3′ ends (Figure 4D). Libraries in which the small RNAs were not subjected to oxidation but were treated with RNA 5′ polyphosphatase displayed strong enrichment of small RNAs with features characteristic of 22G-RNAs: predominant length of 22 nts, strong bias for a 5′G, and production antisense to pos-1 mRNA (Figures 4B and 4C). Oxidation of small RNAs treated with RNA 5′ polyphosphatase caused a depletion in these presumptive secondary siRNAs derived from pos-1 (Figure 4D). Furthermore, the length, 5′ nt distribution, and strand polarity of the small RNAs in this library resembled that of primary siRNAs, indicating that the more abundant 22G-RNAs were depleted and, hence, largely unmethylated (Figures 4B and 4C). In parallel, we assessed whether primary siRNAs are specifically modified in the germline or if somatic siRNAs are also modified. In glp-4(bn2) animals, which fail to undergo germline proliferation when grown at 25°C (Beanan and Strome, 1992), we did not capture small RNAs aligning to pos-1 that resembled 22G-RNAs (i.e., 22-nt-long reads containing a 5′G) regardless of whether the small RNAs were treated with RNA 5′ polyphosphatase (Figure 4E). This is likely because pos-1 is expressed in the germline and, as a consequence, germline-less glp-4 mutants lack an mRNA template for 22G-RNA production. Primary siRNAs, which are derived from the dsRNA administered to the animals and, therefore, not dependent on an endogenous mRNA template, were strongly enriched by oxidation, suggesting that at least a subset of somatic primary siRNAs is also modified (Figures 4D).
We next assessed whether the 3′ modification that we identified on primary siRNAs is dependent on henn-1 by sequencing small RNAs from wild-type and henn-1(pk2295) mutant animals undergoing pos-1 RNAi. As before, the small RNAs were split into oxidized and non-oxidized fractions; however, each fraction was treated with RNA 5′ polyphosphatase and, thus, both primary and secondary siRNAs were captured. Nonetheless, we could distinguish primary siRNAs from secondary siRNAs by their polarity, as any sense-strand siRNAs are presumably produced from the dsRNA administered to the animals and are, thus, primary siRNAs. In wild-type animals, sense strand siRNAs were enriched ~5-fold in the library from oxidized small RNAs relative to the non-oxidized counterpart, consistent with our earlier conclusion that primary siRNAs are modified (Figure S4). In henn-1(pk2295) mutants, however, there was no discernable difference between primary siRNA levels in the oxidized and non-oxidized libraries, indicating that the 3′ modification on these small RNAs is lost in henn-1 mutants (Figure S4).
henn-1 promotes germline RNAi for reasons unknown, although its role in somatic RNAi is confounded by its effect on the ERGO-1-class 26G-RNA pathway, which, when disrupted, enhances RNAi and could, thus, counter modest RNAi defects (Billi et al., 2012; Kamminga et al., 2012; Yigit et al., 2006). Our results indicating that exogenous primary siRNAs are methylated may explain why henn-1 mutants are partially RNAi-defective in the germline. It is possible, however, that the reduction in sid-1 and rde-11 mRNA levels we identified by RNA-seq also contributes to the reduced RNAi sensitivity of henn-1 mutants, as these genes are also required for optimal RNAi (Winston et al., 2002; Yang et al., 2012; Zhang et al., 2012). Our results also demonstrate that each class of small RNAs that acts upstream of the WAGO-class 22G-RNA pathway contains a henn-1-dependent 3′ modification. Because HENN-1 is a 3′ methyltransferase, these results suggest that methylation is uniquely associated with primary small RNAs.
Subpopulations of miRNAs Contain 3′-End Modifications
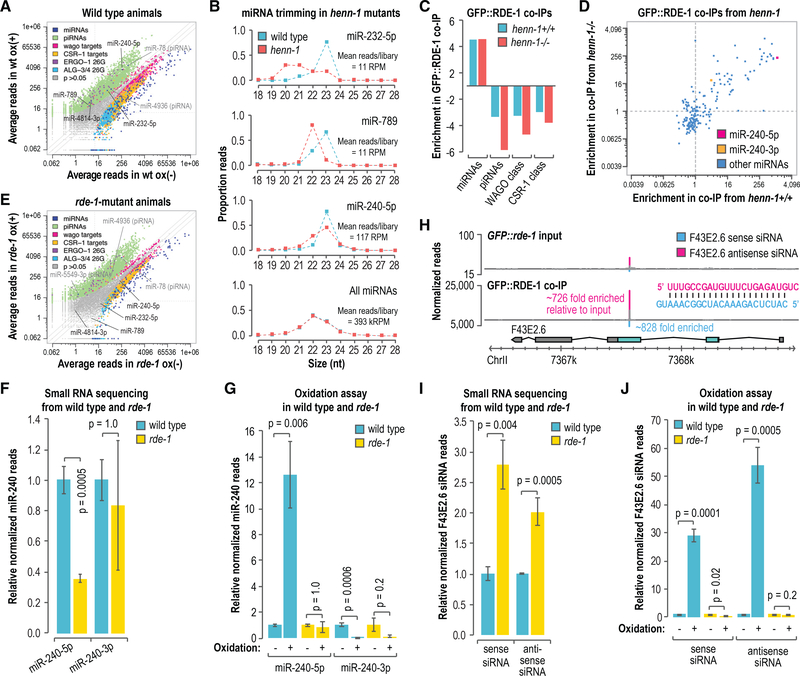
In addition to binding primary siRNAs during RNAi, RDE-1 interacts with miRNAs (Corrêa et al., 2010). miRNAs are not generally methylated in C. elegans (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012), but given our data suggesting that primary siRNAs, which bind RDE-1, are methylated, we tested whether a subset of miRNAs, particularly those bound by RDE-1, might also be methylated. To individually assess miRNA 3′-end modifications, we sequenced small RNAs from wild-type animals after treating the RNA with the oxidizing agent sodium periodate or a control. Consistent with previous studies, piRNAs and ERGO-1-class 26G-RNAs were enriched in libraries prepared from oxidized small RNAs, indicating that they are modified at their 3′ ends (Figure 5A) (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012; Ruby et al., 2006; Vasale et al., 2010). Other classes of small RNAs were largely depleted in the oxidized libraries, although a subset of WAGO-class 22G-RNAs was also modestly enriched, which we did not explore further (Figure 5A). Six small RNAs annotated as miRNAs were enriched in libraries from oxidized small RNAs (Figure 5A). Upon closer inspection, two of these small RNAs, miR-78 and miR-4936, bear hallmarks of piRNAs—21 nt long, contain a 5′ U, and are depleted in prg-1 mutants (Phillips et al., 2015)—suggesting that they are misannotated. Hence, we identified four high-confidence miRNAs, namely, miR-232–5p, miR-240–5p, miR-789, and miR-4814–3p, that were enriched by oxidation and are, therefore, modified at their 3′ ends. Each of these miRNAs was previously shown to interact with RDE-1 (Corrêa et al., 2010). Three of the four miRNAs enriched by oxidation in wild-type animals were truncated in henn-1 mutants, indicating that henn-1 protects them from 3′ trimming and is, thus, likely responsible for modifying their 3′ ends (Figure 5B). The levels of the fourth miRNA miR-4814–3p did not pass our read threshold (10 reads per million total) in our wild-type and henn-1(pk2295) set of libraries and was not analyzed. These data suggest that a subset of miRNAs is modified at their 3′ ends, and because this modification requires henn-1, we conclude that they are likely methylated.
Figure 5. Accumulation of Modified miRNAs and Primary siRNAs Is Dependent on rde-1.
(A) Scatterplot displaying each small RNA feature (miRNA, piRNA, WAGO-class 22G-RNA locus, CSR-1-class 22G-RNA locus, ALG-3/4-class 26G-RNA locus, and ERGO-1-class 26G-RNA locus) as the average number of normalized reads in control (x axis, ox–) and oxidized small RNA libraries (y axis, ox+) (n = 3 biological replicates). Data are from wild-type L4-stage animals.
(B) The proportions of total reads for each miRNA in L4 larvae are plotted by size. The “all miRNAs” plot corresponds to the combined proportions of reads for all annotated miRNAs. The data do not contain biological replicates.
(C) Enrichment or depletion of the indicated classes of small RNAs in GFP::FLAG::RDE-1 coIPs relative to cell lysates. Data are shown for 1 of 2 biological replicates.
(D) Scatterplot displaying each miRNA as its enrichment in GFP::FLAG::RDE-1 coIPs relative to cell lysates from adult animals wild type (henn-1+/+) (x axis) or mutant for henn-1 (henn-1(pk2295)) (y axis). Dashed lines are at 1-fold enrichment on each axis.
(E) Scatterplot displaying each small RNA feature, as in (A), as the average number of normalized reads in control (x axis, ox–) and oxidized small libraries (y axis, ox+) (n = 3 biological replicates). Data are from rde-1(ne219) L4-stage animals grown in parallel to the wild-type animals used in (A).
(F) miR-240–5p and miR-240–3p reads in rde-1(ne219) relative to wild type (n = 3 biological replicates). Error bars report standard deviation.
(G) miR-240–5p and miR-240–3p reads in wild type and rde-1(ne219) from small RNA libraries treated with sodium periodate (oxidation +) relative to libraries receiving a control treatment (oxidation –) (n = 3 biological replicates). Error bars report standard deviation.
(H) Read distribution across the F43E2.6 siRNA-generating locus from GFP::FLAG::rde-1 cell lysate (input, top plot) and GFP::FLAG::RDE-1 coIP small RNA libraries (bottom plot). The most abundant siRNA duplex produced from the locus is shown. Enrichment in the GFP::FLAG::RDE-1 coIP relative to the input is shown for each siRNA.
(I) F43E2.6 sense and antisense siRNA reads in rde-1(ne219) relative to wild type (n = 3 biological replicates). Error bars report standard deviation.
(J) F43E2.6 sense and antisense siRNA reads in wild type and rde-1(ne219) from small RNA libraries treated with sodium periodate (oxidation +) relative to libraries receiving a control treatment (oxidation –) (n = 3 biological replicates). Error bars report standard deviation.
See also Figure S5.
miRNA and Primary siRNA 3′ Modifications Are Dependent on rde-1
To determine if RDE-1 could have a role in promoting small RNA methylation or, alternatively, if small RNA methylation might promote RDE-1 association, we first coIP’d RDE-1 from a strain in which a GFP::FLAG cassette was integrated into the endogenous rde-1 locus using CRISPR-Cas9. Consistent with a previous study in which RDE-1 was coIP’d from an overexpression line, GFP::FLAG::RDE-1 predominantly associated with miRNAs (Figure 5C) (Corrêa et al., 2010). miRNA association with RDE-1 was not dependent on methylation, as GFP::FLAG::RDE-1 largely bound the same miRNAs in both wild-type and henn-1(pk2295) mutants (Figure 5D).
We next tested whether miRNA 3′ modifications are dependent on rde-1. In parallel to the wild-type animals described above (Figure 5A), we sequenced oxidized and non-oxidized small RNAs from rde-1(ne219) mutants. Unlike in wild-type animals, we did not identify any authentic miRNAs that were enriched by oxidation in rde-1(ne219) mutants; however, piRNAs, which also contain a henn-1-dependent 3′ modification were enriched, indicating that the loss of miRNA modifications is not due to henn-1 dysfunction in rde-1 mutants (Figure 5E). Of the four miRNAs enriched by oxidation in our wild-type small RNA sequencing libraries, miR-240–5p was the most strongly enriched in our GFP::FLAG::RDE-1 coIP from henn-1+/+ animals (~1,226-fold) (Figure 5D). miR-240–5p levels were reduced by ~70% in rde-1 mutants (p = 0.0005), which suggests that RDE-1 is its primary binding partner and that it is destabilized in its absence (Figure 5F). In wild-type libraries prepared from oxidized small RNAs, miR-240–5p was enriched ~12-fold relative to non-oxidized small RNA libraries (p = 0.006) (Figure 5G). In contrast, miR-240–5p was not enriched by oxidation in rde-1 mutant libraries (p = 1.0) (Figure 5G). miR-240–3p, which is produced from the same precursor as miR-240–5p but from the opposite arm of the miRNA duplex, was only modestly enriched in our GFP::FLAG::RDE-1 coIP from henn-1+/+ animals (~4-fold) (Figure 5D). We were not able to detect a difference in miR-240–3p levels in rde-1 mutants (Figure 5F). miR-240–3p was not enriched in oxidized small RNA libraries from either wild-type or rde-1 mutants, indicating that despite being processed from the same duplex as miR-240–5p, it is not modified at its 3′ end (Figure 5G). Thus, miR-240–5p, a presumably strong RDE-1 interactor, is enriched by oxidation and, therefore, contains a 3′ modification, whereas miR-240–3p, a relatively weak RDE-1 interactor, is not modified. miR-240–3p also associates with ALG-1 and ALG-2 and miR-240–5p associates with ALG-5, suggesting that the fractions of miR-240 that we detected in rde-1 mutants are likely associated with other Argonautes in an unmethylated form (Brown et al., 2017).
Endogenous canonical siRNAs resembling those produced from exogenous dsRNA are also predicted to bind RDE-1. Indeed, we identified endogenous siRNAs produced from the F43E2.6 gene locus that were strongly enriched in our GFP::FLAG::RDE-1 coIPs (Figure 5H). F43E2.6 siRNAs were produced from a duplex that is 23 nt long and has 2-nt 3′ overhangs, indicating that it is likely processed by Dicer and is, therefore, a canonical siRNA locus (Figure 5H) (Ameres and Zamore, 2013; Warf et al., 2012). Furthermore, the levels of F43E2.6 siRNAs were elevated in mut-16 mutants, indicating that they are not produced through the Mutator pathway, unlike WAGO-class 22G-RNAs (Figure S5A) (Phillips et al., 2012; Zhang et al., 2011). F43E2.6 siRNAs were modestly elevated in rde-1 mutants (p ≤ 0.004), suggesting that association with RDE-1 destabilizes them relative to other Argonautes they might interact with or that their production is elevated in the absence of rde-1 (Figure 5I). Both sense and antisense F43E2.6 siRNAs were enriched in GFP::FLAG::ALG-1 coIPs, indicating that they are partitioned between at least two Argonautes (Figure S5B) (Brown et al., 2017). In wild-type samples, the F43E2.6 sense strand siRNA was enriched by ~30-fold and the antisense siRNA by ~55-fold in oxidized small RNA libraries relative to non-oxidized small libraries, indicating that both strands of the duplex contain a 3′ modification (p ≤ 0.0005) (Figure 5J). Conversely, neither siRNA was enriched in oxidized small RNA libraries from rde-1 mutants, suggesting that accumulation of these siRNAs in their 3′ modified form is dependent on their association with RDE-1 (Figure 5J). These results point to a link between RDE-1 association and miRNA and primary siRNA 3′ modification, a modification dependent on henn-1 and, therefore, likely methylation, and thus, we speculate that RDE-1 may direct small RNA methylation.
DISCUSSION
The germline mortality of henn-1 mutants likely relates to its role in the piRNA pathway, as disrupting the other small RNA pathways that henn-1 is implicated in, those involving the Argonautes ERGO-1 and RDE-1, does not result in progressive sterility (Figures S1C and S1D) (Sakaguchi et al., 2014). The progressive sterility of henn-1 mutants follows a similar, albeit temperature sensitive, progression to that of prg-1/piwi mutants (Simon et al., 2014). Immediate sterility ensues after reestablishing the WAGO pathway, and therefore endogenous RNAi, in the absence of maternal henn-1 or prg-1 (de Albuquerque et al., 2015; Phillips et al., 2015). Animals heterozygous for henn-1 in which the wild-type copy of the gene was contributed by the father, which presumably becomes active at the maternal-to-zygotic transition that occurs at the ~28-cell stage, displayed equal rates of sterility to animals homozygous mutant for henn-1 after reestablishing endogenous RNAi. Thus, a model emerges in which maternally inherited methylated piRNAs and/or henn-1 establish proper RNA silencing during embryogenesis, which presumably persists throughout germline development to ultimately impact fertility. This would likely occur through the initiation of secondary WAGO-class 22G-RNA production, which is thought to provide a memory of piRNA activity (Ashe et al., 2012; Buckley et al., 2012; Luteijn et al., 2012; Shirayama et al., 2012). In support of this, we observed similar effects on 22G-RNA levels across development in henn-1 mutants, despite henn-1 only being required for piRNA stability in embryos and early larval stages (Billi et al., 2012).
Although henn-1, and by extension methylation, has an important role in germline proliferation, it appears to have little impact on piRNA stability in the adult germline. Why methylation is important for piRNA stability in embryos but not in adult germ cells is unclear. It is possible that the genes involved in piRNA decay in embryos are absent in adults; however, we saw a similar degree of piRNA trimming in wild-type embryos and adults, suggesting there may be two distinct mechanisms for piRNA decay, only one of which is affected by methylation. Alternatively, methylation may be specifically required for stabilizing piRNAs during maternal deposition in the embryo. P granules, the germ granules that house PRG-1/Piwi, break away from nuclear envelope after fertilization, possibly releasing piRNAs into the cytoplasm, which may expose them to decay machinery that they are normally protected from in P granules (Seydoux, 2018). It is also possible that piRNAs engage a distinct set of targets in embryos to which they have extensive complementarity at their 3′ ends and as a consequence are more susceptible to target-directed decay. This would hint at a role for piRNAs in early development that is perhaps distinct from their role later in development. piRNA-mRNA interactions that require only partial complementarity, which is common in C. elegans (Shen et al., 2018; Zhang et al., 2018), may not be affected by methylation. Modulation of methylation may even promote one function of piRNAs over another, which could explain why genes misregulated in henn-1 mutants, despite nearly all being potential piRNA targets, were not uniformly up- or downregulated.
Our results demonstrate that small RNAs that act upstream of the Mutator complex, the WAGO-class 22G-RNA amplification component of the RNAi pathway, contain a 3′ modification that is dependent on henn-1 and, thus, presumably methylation, whereas those that do not act as primary small RNAs are not modified (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012). The Mutator complex contains a nucleotidyltransferase (MUT-2) and a 3′–5′ exonuclease (MUT-7), both of which could promote decay of primary small RNAs that feed into the pathway (Phillips et al., 2012). It is possible that methylation protects primary small RNAs from degradation by the RNAi machinery or by factors that bridge the primary and secondary small RNA pathways. In support of this model, in Arabidopsis, methylation protects small RNAs from uridylation by HESO1, which is recruited to mRNA targets by the Argonaute AGO1 to facilitate decay of the 5′ fragment generated by AGO1 cleavage (Wang et al., 2018).
We showed that rde-1 is required for the accumulation of miRNAs and canonical siRNAs containing modifications at their 3′ ends. Although we identified only a small subset of miRNAs that were resistant to oxidation, an indicator of the presence of a 3′ modification, most miRNAs were enriched in our RDE-1 coIPs. Therefore, if RDE-1 directs small RNA methylation, subpopulations of most individual miRNAs are likely methylated. Methylated miRNAs bound to RDE-1 may have a distinct function from non-methylated miRNAs bound to the other miRNA-associated Argonautes, ALG-1, ALG-2, and ALG-5. miR-243, for example, promotes mRNA entry into the endogenous RNAi pathway through its association with RDE-1 (Corrêa et al., 2010). miR-243, however, was not enriched by oxidation possibly because it also binds strongly to ALG-1 (Brown et al., 2017). In Drosophila, miRNAs predominantly associate with Ago1 and are not methylated; however, some also associate with the predominantly siRNA-associated Argonaute Ago2 and are methylated, consistent with Drosophila Ago2 directing small RNA methylation (Ameres et al., 2010; Horwich et al., 2007). It seems an analogous system may exist in C. elegans between the miRNA-associated Argonaute ALG-1, ALG-2, and ALG-5 and the exogenous siRNA-Argonaute RDE-1. It was recently shown that miRNAs are methylated in sea anemones, which commonly interact with near-perfect complementarity to their targets (Modepalli et al., 2018). Together with these studies, our results raise the possibility that some degree of miRNA methylation is common in animals.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT AND MATERIALS AVAILABILITY
Requests for information and resources should be directed to and will be fulfilled by the lead contact, Taiowa Montgomery (tai. montgomery@colostate.edu). All C. elegans strains generated in this study are available on request from the Lead Contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The Key Resources Table contains all of the C. elegans strains used in this study. Hermaphroditic animals were grown on nematode growth medium and fed E. coli OP50, except where noted otherwise. Animals used in the experiments in Figures 1A, 1B, 1F, 1G, 4, S1A–S1C, S1E, and S4 were grown at 25°C. All other animals were grown at 20°C. The developmental stage at which the animals were analyzed is specified in the methods and figures.
METHOD DETAILS
Strain Generation
USC1080[rde-1(cmp133[(gfp + loxP + 3xFLAG)::rde-1]) V] and USC988[mut-16(cmp41[mut-16::mCherry + loxP]) wago-1(cmp92 [(GFP + loxP + 3xFLAG)::wago-1]) I] were generated by introducing a GFP::3xFLAG cassette at the 5′ end of the endogenous rde-1 or wago-1 coding sequence using CRISPR-Cas9 genome editing (Arribere et al., 2014; Dickinson et al., 2013, 2015). The repair templates were amplified using primers listed in Table S5 and introduced into the vector pDD282 (Addgene #66823) by isothermal assembly (Gibson et al., 2009). To protect the repair template from cleavage, a mutation of the PAM sequence was incorporated into one of the primers. The guide RNA plasmid was generated by ligating oligos containing the guide RNA sequence into BsaI-digested pRB1017 (Addgene #59936) (Dickinson et al., 2013). TAM18 [henn-1(ram13[henn-1ko; mCherry::Flag-no-stop-codon]) III] was also generated using CRISPR-Cas9 (Dickinson et al., 2013). The repair template was amplified with the primers listed in Table S5 and introduced into SpeI and ClaI digested pJJR83 (Addgene #75028). The guide RNA construct was generated with the NEBuilder kit (New England BioLabs, E2621) using the plasmid pDD162 (Addgene #47549) and the primers listed in the Table S5. The entire henn-1 coding sequence was replaced with an mCherry::3xFLAG cassette lacking a stop codon and thus does not express mCherry. TAM226[mut-16(pk710) I; henn-1(pk2295) III], TAM236[mut-16(pk710) I; henn-1(ram13) III], TAM27[henn-1(pk2295) III; mut-14(mg464) smut-1(tm1301) V], and TAM35[henn-1(ram13) III; mut-14(mg464) smut-1(tm1301) V] were generated using standard genetic methods.
Multigenerational Brood Size Assays
Worms were grown on a continuous supply of E. coli (OP50) and chunked to fresh plates at each new generation (every 2–3 days). Because animals lay eggs over multiple days, the number of generations is approximate. Brood size assays were performed by separating 15 L4 larvae per strain onto individual plates, removing their offspring twice per day, and recording the number removed until the animals were no longer laying eggs. P values were calculated using Wilcoxon rank-sum tests.
Transgenerational Sterility Assays
Transgenerational sterility assays were modeled after the approach used by Simon et al. (2014) and Ahmed and Hodgkin (2000). Briefly, 10 independent lines per wild-type, henn-1(pk2295), prg-1(n4357), and ergo-1(tm1860) strain were grown at 20°C or 25°C. 10 worms per line were transferred to new plates every generation (2–3 days at 25°C or 3–4 days at 20°C) for 30 generations, and fertility was recorded at each transfer. Animals producing any number of offspring were considered fertile, and all offspring were transferred from plates on which there were fewer than 10 viable progeny.
Reestablishing Endogenous RNAi
The RNAi pathway was restored in RNAi-defective mutants using the genetics approach illustrated in Figure 1C as described (Phillips et al., 2015). Individual F1 animals from each cross were genotyped to confirm heterozygosity of smut-1(tm1301), indicating that the progeny were from a cross. Animals that were wild-type or homozygous mutant for smut-1 were excluded from the analysis. P values were calculated using two-sample t tests.
Imaging
Animals were imaged on a Zeiss Axio Imager Z2 upright microscope after immobilization in 10–25 uM sodium azide on 1.5% Agarose pads on glass slides.
Gonad Dissections
Animals were grown to gravid adult stage at 20°C for 68–70 hours after L1 synchronization and dissected by cutting posterior to the pharynx or anterior to the tail in egg buffer containing 10 mM levamisole. Extruded distal gonads were collected by mouth pipetting, transferred to 500 ul Trizol, and frozen at −80°C (Campbell and Updike, 2015). Because the gonads break at the arms, the samples did not include the proximal gonad arms, which contain mature oocytes and sperm.
RNA Isolation
Dissected gonads and whole animals were flash frozen in liquid nitrogen prior to RNA isolation. RNA was isolated using Trizol (Life Technologies) followed by two chloroform extractions. RNA was precipitated in isopropanol for 20 minutes at room temperature for whole animal samples and overnight at −80°C in the presence of 20 ug glycogen for germline, input, and co-IP samples.
Quantitative Real-Time PCR
Quantitative real-time-PCR of small RNAs was done as described using TaqMan reagents and custom probes following the manufacturers recommendations (Billi et al., 2012; Montgomery et al., 2012). Ct values were captured using a Bio-Rad CF96 Real-Time PCR Detection System. Ct values were averaged across three technical replicates. Mean relative 21U-RNA levels across 3 biological replicates were calculated using the 2−ddCt method (Livak and Schmittgen, 2001). miR-35 levels were used for normalization. Two-sample t tests were used to compare differences between samples. Bonferroni adjustments were applied to correct for multiple comparisons.
Small RNA Northern Blot
10 ug total RNA from each sample was resolved on a 17% denaturing polyacrylamide gel, transferred to a Hybond-N+ membrane (GE Healthcare), and UV crosslinked. T4 polynucleotide kinase (NEB) was used to radiolabel an LNA oligonucleotide antisense to 21UR-1 with 32P-ATP. The membrane was incubated with the probe overnight at 38°C in PerfectHyb Plus Hybridization Buffer (Sigma) and washed 5 times in SSC/SDS buffer at 50°C for 20 minutes each wash.
Small RNA-Seq Libraries
Small RNAs were size selected (~16–30 nt) on 17% polyacrylamide/urea gels. Small RNA treatment varied by library as indicated in Table S6. Oxidation to enrich for 3′-2′-O-methylated small RNAs was done using 200 mM sodium periodate in borate buffer (pH 8.6) as described, but without beta-elimination (Montgomery et al., 2012; Yu and Chen, 2010). Polyphosphatase treatment to reduce 5′ di- and triphosphates to monophosphates was done using RNA 5′ polyphosphatase (Illumina). High-throughput sequencing libraries were prepared as described (Brown et al., 2017) or using the NEBNext Multiplex Small RNA Library Prep Kit (Illumina) (Table S6). The 3′ ligation was done at 16°C for 18 hours to improve capture of methylated small RNAs. PCR amplicons were size selected on 10% polyacrylamide non-denaturing gels.
Small RNA-Seq Data Analysis
16–30 nt small RNA sequences were parsed from the library adapters, filtered to exclude reads with >1 base having a Phred quality score < 30 and mapped to the C. elegans genome (Wormbase release WS230) using CASHX v. 2.3 (Fahlgren et al., 2009). miRNAs, piRNAs, WAGO-class 22G-RNA loci, CSR-1-class 22G-RNA loci, ALG-3/4-class 26G-RNA loci, and ERGO-1-class 26G-RNA loci were classified as described (Phillips et al., 2015). Differential expression analysis, including fold change and p value calculations, was done using DESeq2 v. 1.18.1 with the Benjamin Hochberg multiple test correction applied (FDR = 0.05) (Love et al., 2014). All other p values reported for small RNA sequencing data were calculated using two-sample t tests. Custom Perl and Python scripts, R, and Excel were used for all other data analyses. Plots were drawn in R, Excel, BioVenn, and IGV (Hulsen et al., 2008; Thorvaldsdóttir et al., 2013). Sequencing was done on an Illumina NextSeq 500 (High Output Kit, 75 cycles) at the Colorado State University Next-Gen Sequencing Core.
RNA-Seq Libraries
Ribosomal RNA was depleted from total RNA using Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) (Illumina). rRNA depleted RNA was purified using the RNA Clean & Concentrator kit (Zymo Research). DNase treatment was done on the columns used for RNA purification. High-throughput sequencing libraries were prepared from purified rRNA-depleted RNA using NEBNext Ultra II Directional RNA Library Prep Kit (NEB). Most sequencing was done on an Illumina NextSeq 500 (High Output Kit, 75 cycles) at the Colorado State University Next-Gen Sequencing Core. (Table S6).
RNA-Seq Data Analysis
Adapters and low-quality bases were removed using Trimmomatic v. 0.35 (Bolger et al., 2014). Reads were mapped to the C. elegans genome (Wormbase release WS230) using Star v. 2.5.0a (Dobin et al., 2013). Reads from specific genes were counted using RSEM v. 1.3.0 (Li and Dewey, 2011). Differential expression analysis, including fold change and p value calculation, was done using DESeq2 v. 1.18.1 with the Benjamin Hochberg multiple test correction applied (FDR = 0.05) (Love et al., 2014). Plots were drawn with R and IGV (Thorvaldsdóttir et al., 2013).
Transposon Analysis
To identify reads mapping to each annotated transposon family, mRNA and small RNA sequencing reads were aligned to each of 152 transposon family consensus sequences using Bowtie (Langmead, 2010; Jurka, 2000). Reads mapping to more than one transposon family were discarded. Differential expression analysis, including fold change and p value calculation, was done using DESeq2 v. 1.18.1 (Love et al., 2014).
RNAi Assays
Synchronized L1 larvae were transferred to plates with E. coli HT115 containing an empty vector (L4440) or expressing dsRNA complementary to pos-1 (Kamath et al., 2003) and grown at 25°C. Gravid adults were collected for RNA isolation at 48 hours.
Co-Immunoprecipitations
FLAG::HRDE-1, GFP::FLAG::WAGO-1, and GFP::FLAG::RDE-1 were coimmunoprecipitated from gravid adults 68 hours after L1 synchronization. Animals were flash frozen in liquid nitrogen and lysed in 50 mM Tris-Cl, pH 7.4, 100 mM KCl, 2.5 mM MgCl2, 0.1% Igepal CA-630, 0.5 mM PMSF, and 1X Pierce Protease Inhibitor Tablets. Cell lysates were cleared by centrifugation for 10 min at 12,000 RCF and incubated with anti-GFP mAb-agarose for 1 hour or with anti-FLAG followed by Protein G magnetic beads for a combination of 1 hour. After co-immunoprecipitation, beads were washed in lysis buffer and split into RNA and protein fractions. All steps were done at 4°C. Antibody specificity for FLAG (FLAG::HRDE-1, GFP::FLAG::WAGO-1 co-IPs) or GFP (GFP::FLAG::RDE-1) was confirmed by doing negative control co-IPs from wild-type animals.
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical tests used are described within the methods section describing each experiment. Bonferroni multiple test corrections were applied to p values when comparing >2 conditions.
DATA AND CODE AVAILABILITY
All the high-throughput sequencing data described here has been deposited to the Gene Expression Omnibus (GEO) and is freely available under accession number GEO: GSE137734. Software used in the study is described in the corresponding methods section and the Key Resources Table. Custom R, Perl, and Python scripts are freely available on request.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GFP mAb-agarose | MBL | Cat# D153–8; RRID:AB_591815 |
| anti-FLAG | Sigma | Cat# F1804; RRID:AB_262044 |
| Bacterial Strains | ||
| E. coli OP50 | Gary Ruvkun (Harvard Medical School) | N/A |
| E. coli HT115 | Gary Ruvkun (Harvard Medical School) (Kamath et al., 2003) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trizol | Life Technologies | 15596018 |
| RNA 5′ polyphosphatase | Illumina | RP8092H |
| NEBNext 2X PCR Master Mix | NEB | M0541L |
| TURBO DNase | Life Technologies | AM2238 |
| AMPure XP Beads | Beckman Coulter | A63881 |
| Pierce Protease Inhibitor Tablets | Pierce Biotechnology | 88266 |
| Amersham Hybond -N+ Membranes | GE Healthcare | RPN303B |
| PerfectHyb Plus Hybridization Buffer | Sigma-Aldrich | H7033 |
| T4 Polynucleotide Kinase (3′ phosphatase minus) | NEB | M0236S |
| SureBeads Protein G Magnetic Beads | Bio-Rad | 161–4023 |
| Critical Commercial Assays | ||
| TaqMan | Life Technologies | 4366596, 4444557, 4427975 |
| NEBNext Ultra II Directional RNA Library Prep Kit for Illumina | NEB | E7760S |
| RNA Clean & Concentrator | Zymo Research | R1015 |
| Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) | Illumina | MRZH11124 |
| NEBNext Small RNA Library Prep Kit for Illumina | NEB | E7300S or E7580S |
| Deposited Data | ||
| NGS data | This Study | Gene Expression Omnibus GSE137734 |
| Experimental Models: Organisms/Strains | ||
| C. elegans Bristol Strain | CGC | N2 |
| glp-4(bn2) I | CGC | SS104 |
| henn-1(pk2295) III | René Ketting (IMB Mainz) | NL4415 |
| prg-1(n4357) I | CGC | SX922 |
| rde-1(ne219) V | CGC | WM27 |
| mut-14(mg464) smut-1(tm1301) V | Phillips et al., 2015 | GR1948 |
| mut-16(pk710) I | CGC | NL1810 |
| henn-1(pk2452) III | René Ketting (IMB Mainz) | RFK30 |
| glh-1(sam24[glh-1::gfp::3Xflag]) I | Dustin Updike (MDI) | DUP64 |
| glh-1(sam24[glh-1::gfp::3Xflag]) I; henn-1(pk2295) III | This Study | TAM56 |
| rde-1(cmp133[(gfp + loxP + 3xFLAG)::rde-1]) V | This Study | USC1080 |
| mut-16(cmp41[mut-16::mCherry + loxP]) wago-1(cmp92[(GFP + loxP + 3xFLAG)::wago-1]) I | This Study | USC988 |
| henn-1(ram13[henn-1ko; mCherry::Flag-no-stop-codon]) III | This Study | TAM18 |
| mut-16(pk710) I; henn-1(pk2295) III | This Study | TAM26 |
| mut-16(pk710) I; henn-1(ram13) III | This Study | TAM36 |
| henn-1(pk2295) III; mut-14(mg464) smut-1(tm1301) V | This Study | TAM27 |
| henn-1(ram13) III; mut-14(mg464) smut-1(tm1301) V | This Study | TAM35 |
| flag::hrde-1([flag::hrde-1, cb-unc-119(+)]) II; unc-119(ed3) III | Craig Mello (University of Massachusetts) (Shirayama et al., 2012) | N/A |
| Oligonucleotides | ||
| See Table S5 | See Table S5 | N/A |
| Recombinant DNA | ||
| pDD282 (GFP::3xFLAG repair template plasmid) | Addgene (Dickinson et al., 2015) | 66823 |
| pRB1017 (Guide RNA plasmid) | Addgene (Arribere et al., 2014) | 59936 |
| pJJR83 (mCherry::3xFLAG repair template plasmid) | Addgene (a gift from Mike Boxem) | 75028 |
| pDD162 (Guide RNA plasmid) | Addgene (Dickinson et al., 2013) | 47549 |
| L4440 (RNAi vector) | Addgene (a gift from Andrew Fire) | 1654 |
| pos-1 RNAi plasmid | Gary Ruvkun (Harvard Medical School) (Kamath et al., 2003) | N/A |
| Software and Algorithms | ||
| CASHX v. 2.3 | Fahlgren et al., 2009 | N/A |
| DESeq2 v. 1.18.1 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Trimmomatic v. 0.35 | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Star v. 2.5.0a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Bowtie | Langmead, 2010 | http://bowtie-bio.sourceforge.net/index.shtml |
| RSEM v. 1.3.0 | Li and Dewey, 2011 | https://deweylab.github.io/RSEM/ |
| Zen Image Analysis | Zeiss | N/A |
Highlights.
henn-1 mutants display transgenerational sterility
Maternal henn-1 is required to establish proper RNA silencing
HENN-1 stabilizes piRNAs in embryos but is dispensable in the adult germline
RDE-1-associated siRNAs and miRNAs are modified at their 3′ ends by HENN-1
ACKNOWLEDGMENTS
Thanks to Mark Stenglein, Justin Lee, and Marylee Layton for assistance with sequencing; Dustin Updike for training in gonad dissections and for providing the glh-1::GFP strain; and Rene Ketting for providing henn-1(pk2452). Additional strains were provided by the CGC, which is funded by the NIH (P40 OD010440). This work was supported by the Boettcher Foundation (003614–00002 to T.A.M.), the NIH (R35GM119775 to T.A.M., T32GM132057 to K.J.R., R35GM119656 to C.M.P., and T32GM118289 to D.H.N.), and Pew Charitable Trusts (C.M.P.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.10.114.
REFERENCES
- Ahmed S, and Hodgkin J (2000). MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403, 159–164. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, and Jewell D (2003). MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13, 807–818. [DOI] [PubMed] [Google Scholar]
- Ameres SL, and Zamore PD (2013). Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475–488. [DOI] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, and Zamore PD (2010). Target RNA-directed trimming and tailing of small silencing RNAs. Science 328, 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andralojc KM, Campbell AC, Kelly AL, Terrey M, Tanner PC, Gans IM, Senter-Zapata MJ, Khokhar ES, and Updike DL (2017). ELLI-1, a novel germline protein, modulates RNAi activity and P-granule accumulation in Caenorhabditis elegans. PLoS Genet. 13, e1006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, and Tabara H (2007). In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26, 5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, and Fire AZ (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. (2012). piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, and Miska EA (2012). Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanan MJ, and Strome S (1992). Characterization of a germ-line proliferation mutation in C. elegans. Development 116, 755–766. [DOI] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, and Kim JK (2012). The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 8, e1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Fischer SE, and Kim JK (2014). Endogenous RNAi pathways in C. elegans. WormBook, 1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KC, Svendsen JM, Tucci RM, Montgomery BE, and Montgomery TA (2017). ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline. Nucleic Acids Res. 45, 9093–9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, and Kennedy S (2012). A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AC, and Updike DL (2015). CSR-1 and P granules suppress sperm-specific transcription in the C. elegans germline. Development 142, 1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. (2009). The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa RL, Steiner FA, Berezikov E, and Ketting RF (2010). MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet. 6, e1000903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque BF, Placentino M, and Ketting RF (2015). Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans. Dev. Cell 34, 448–456. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, and Goldstein B (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, and Goldstein B (2015). Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics 200, 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Sullivan CM, Kasschau KD, Chapman EJ, Cumbie JS, Montgomery TA, Gilbert SD, Dasenko M, Backman TW, Givan SA, and Carrington JC (2009). Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA 15, 992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr., Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. (2009). Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36, 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heestand B, Simon M, Frenk S, Titov D, and Ahmed S (2018). Transgenerational Sterility of Piwi Mutants Represents a Dynamic Form of Adult Reproductive Diapause. Cell Rep. 23, 156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, and Zamore PD (2007). The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17, 1265–1272. [DOI] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, and Alkema W (2008). BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J (2000). Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16, 418–420. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, and Ketting RF (2012). Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 8, e1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, and Tomari Y (2010). Making RISC. Trends Biochem. Sci. 35, 368–376. [DOI] [PubMed] [Google Scholar]
- Kurth HM, and Mochizuki K (2009). 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 15, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B (2010). Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics Chapter 11, Unit 11.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D Jr., and Mello CC (2012). C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, and Chen X (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15, 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, and Ketting RF (2012). Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31, 3422–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurchy AN, Stempor P, Gaarenstroom T, Wysolmerski B, Dong Y, Aussianikava D, Appert A, Huang N, Kolasinska-Zwierz P, Sapetschnig A, et al. (2017). A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. eLife 6, e21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modepalli V, Fridrich A, Agron M, and Moran Y (2018). The methyltransferase HEN1 is required in Nematostella vectensis for microRNA and piRNA stability as well as larval metamorphosis. PLoS Genet. 14, e1007590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, and Ruvkun G (2012). PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 8, e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafó DB, and Robb GB (2010). Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA 16, 2537–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, and Zamore PD (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108. [DOI] [PubMed] [Google Scholar]
- Pak J, and Fire A (2007). Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315, 241–244. [DOI] [PubMed] [Google Scholar]
- Pazdernik N, and Schedl T (2013). Introduction to germ cell development in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Montgomery TA, Breen PC, and Ruvkun G (2012). MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Brown KC, Montgomery BE, Ruvkun G, and Montgomery TA (2015). piRNAs and piRNA-Dependent siRNAs Protect Conserved and Essential C. elegans Genes from Misrouting into the RNAi Pathway. Dev. Cell 34, 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, and Bartel DP (2006). Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207. [DOI] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, and Siomi MC (2007). Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 21, 1603–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A, Sarkies P, Simon M, Doebley AL, Goldstein LD, Hedges A, Ikegami K, Alvares SM, Yang L, LaRocque JR, et al. (2014). Caenorhabditis elegans RSD-2 and RSD-6 promote germ cell immortality by maintaining small interfering RNA populations. Proc. Natl. Acad. Sci. USA 111, E4323–E4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, and Zamore PD (2008). Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA* strands in flies. Curr. Biol. 18, 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Gu W, Ishidate T, Conte D Jr., and Mello CC (2013). The C. elegans CSR-1 Argonaute Pathway Counteracts Epigenetic Silencing to Promote Germline Gene Expression. Dev. Cell 27, 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G (2018). The P Granules of C. elegans: A Genetic Model for the Study of RNA-Protein Condensates. J. Mol. Biol. 430, 4702–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EZ, Chen H, Ozturk AR, Tu S, Shirayama M, Tang W, Ding YH, Dai SY, Weng Z, and Mello CC (2018). Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 172, 937–951.e918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D Jr., and Mello CC (2012). piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, and Plasterk RH (2007). Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247. [DOI] [PubMed] [Google Scholar]
- Simon M, Sarkies P, Ikegami K, Doebley AL, Goldstein LD, Mitchell J, Sakaguchi A, Miska EA, and Ahmed S (2014). Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep. 7, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, and Mello CC (2002). The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109, 861–871. [DOI] [PubMed] [Google Scholar]
- Thomson T, and Lin H (2009). The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 25, 355–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, and Conte D Jr. (2010). Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA 107, 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Fields BD, Spracklin G, Shukla A, Phillips CM, and Kennedy S (2018). Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Dou Y, Chen L, Wang J, Jiang N, Guo C, Yao Q, Wang C, Liu L, et al. (2018). Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3′ to 5′ exoribonuclease Atrimmer 2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 115, E6659–E6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Shepherd BA, Johnson WE, and Bass BL (2012). Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res. 22, 1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeles CJ, Wu MZ, and Claycomb JM (2013). Protection of Germline Gene Expression by the C. elegans Argonaute CSR-1. Dev. Cell 27, 664–671. [DOI] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, and Hunter CP (2002). Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459. [DOI] [PubMed] [Google Scholar]
- Wu WS, Brown JS, Chen TT, Chu YH, Huang WC, Tu S, and Lee HC (2019). piRTarBase: a database of piRNA targeting sites and their roles in gene regulation. Nucleic Acids Res. 47, D181–D187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Feng X, Chen X, Weng C, Yan Q, Xu T, Hong M, and Guang S (2018). A Cytoplasmic Argonaute Protein Promotes the Inheritance of RNAi. Cell Rep. 23, 2482–2494. [DOI] [PubMed] [Google Scholar]
- Yang H, Zhang Y, Vallandingham J, Li H, Florens L, and Mak HY (2012). The RDE-10/RDE-11 complex triggers RNAi-induced mRNA degradation by association with target mRNA in C. elegans. Genes Dev. 26, 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, and Mello CC (2006). Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747–757. [DOI] [PubMed] [Google Scholar]
- Yu B, and Chen X (2010). Analysis of miRNA Modifications. Methods Mol. Biol. 592, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, and Chen X (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, and Ruvkun G (2011). mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108, 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Fischer SE, Garcia SM, Riedel CG, Fahlgren N, Sullivan CM, Carrington JC, and Ruvkun G (2012). The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Curr. Biol. 22, 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tu S, Stubna M, Wu WS, Huang WC, Weng Z, and Lee HC (2018). The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359, 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the high-throughput sequencing data described here has been deposited to the Gene Expression Omnibus (GEO) and is freely available under accession number GEO: GSE137734. Software used in the study is described in the corresponding methods section and the Key Resources Table. Custom R, Perl, and Python scripts are freely available on request.