Figure 4. Primary siRNAs Are Modified at Their 3′ Ends.
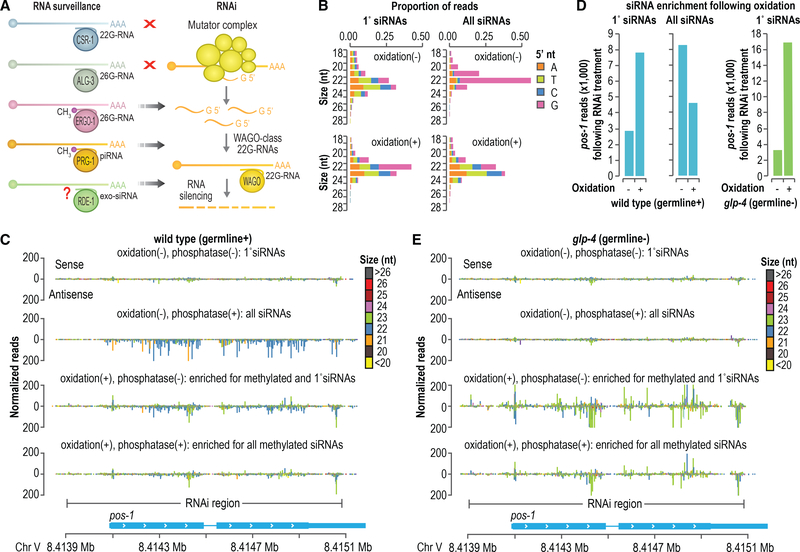
(A) Model depicting the interplay between RNA surveillance and WAGO-class 22G-RNA production by the RNAi pathway.
(B) Size distribution and 5′ nucleotide composition of pos-1 siRNAs following pos-1 RNAi, as determined by high-throughput sequencing. Small RNAs were treated with sodium periodate (oxidation) to enrich for methylated small RNAs or subjected to a control treatment prior to library preparation. The “1° siRNAs” plot corresponds to libraries in which the small RNAs were not treated with RNA 5′ polyphosphatase and, thus, do not include di- and tri-phosphorylated small RNAs (e.g., 22G-RNAs). The “All siRNAs” plot corresponds to libraries in which the small RNAs were treated with RNA 5′ polyphosphatase and, thus, include both 1° and 2° siRNAs.
(C) Small RNA read distribution across pos-1 from wild-type high-throughput sequencing libraries in which the small RNAs were treated with sodium periodate (oxidation +) and RNA 5′ polyphosphatase (polyphosphatase +) as indicated.
(D) Bar plots displaying normalized reads (reads per million total mapped reads) mapping to pos-1 following sodium periodate (oxidation +) or control treatment (oxidation –). The plots on the left are from wild-type animals, and the plot on the right is from glp-4(bn2) mutants, which lack proliferated germlines and for which only primary siRNAs are shown.
(E) Small RNA read distribution across pos-1 from glp-4(bn2) high-throughput sequencing libraries in which the small RNAs were treated with sodium periodate (oxidation +) and RNA polyphosphatase (polyphosphatase +) as indicated.
See also Figure S4.