Figure 3.
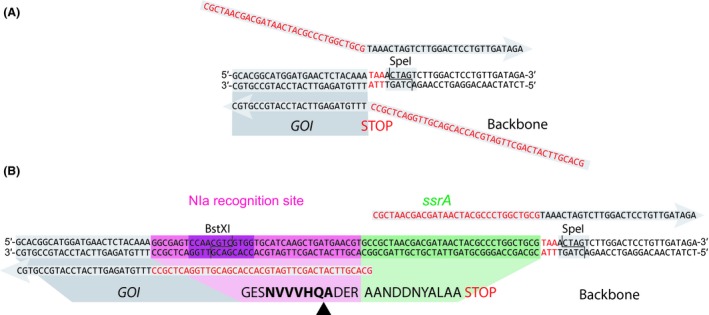
Introduction of the hybrid NIa/SsrA degradation tag into a target protein.A. The degradation tag can be fused to a gene of interest by site‐directed mutagenesis PCR. Primers used to fuse the tag to the monomeric superfolder GFP (msfGFP) in plasmid pS23·Pm‐GFP* are shown in dark grey. The 3′‐terminus of the primers anneals to the plasmid (black letters) and primes amplification of the fragment. The 5′‐ends carry the synthetic NIa/SsrA tag proper (red letters).B. After re‐circularization of the PCR product, the resulting plasmid contains the NIa/SsrA tag fused to the gene of interest. The tag includes the conserved NIa recognition site (indicated in boldface) with its cleavage site (black triangle) followed on the 3′‐terminal by the predicted SsrA degradation signal from P. aeruginosa (Flynn et al., 2001). Alternatively, other cloning strategies can be used to insert the degradation tag for conditional proteolysis, such as restriction/ligation (relevant restriction sites, compatible with the Standard European Vector Architecture, are indicated), or a different cloning technique (e.g. USER or Gibson assembly).
