Figure 2.
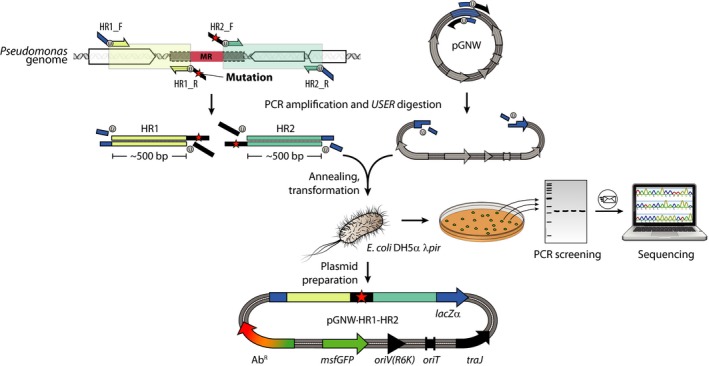
Workflow for the construction of derivatives of pGNW vectors for genome engineering. Two homology regions (HR1 and HR2), each one spanning about 500 bp and located upstream and downstream of the mutagenesis region (MR), are amplified from genomic DNA of Pseudomonas via PCR. Modifications (i.e. insertions or substitutions) are introduced between the HRs as overhangs in the oligonucleotides (indicated in the diagram with a red star) or as additional DNA fragments (not shown). For gene deletions, the sequences of the HRs are designed so that they frame the genome sequence to be deleted. HR1 and HR2 are fused and integrated into one of the pGNW vectors via USER assembly (shown here) or alternative molecular cloning techniques. The thereby assembled pGNW plasmid is then introduced into E. coli DH5α λpir. Individual E. coli clones obtained after transformation are examined for green fluorescence on a blue‐light transilluminator and checked for the correct pGNW insert size via colony PCR, and the purified amplicon is sent for sequencing. After the sequence integrity is confirmed, pGNW plasmids are purified from the respective E. coli strain and saved for the next step.
