Figure 5.
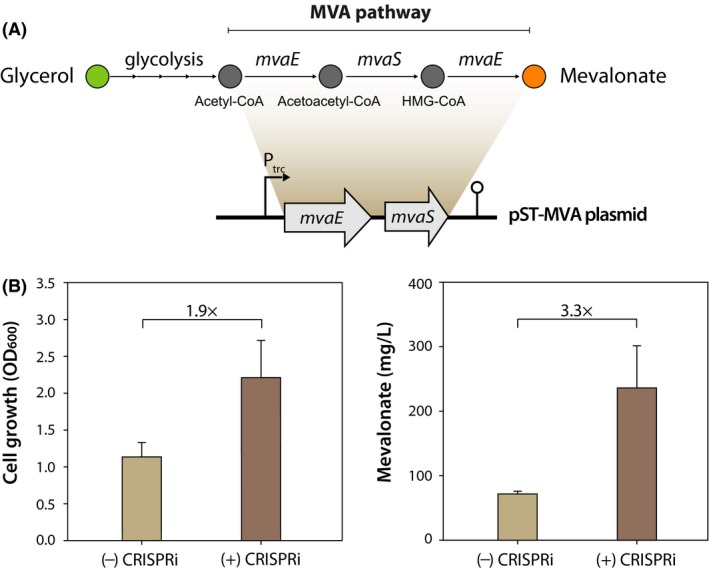
Application of the CRISPRi system for enhancing MVA production in Pseudomonas putida KT2440. (A) Schematic representation of the MVA production pathway and plasmid (pST‐MVA). The engineered MVA pathway encoded by pST‐MVA plasmid consists of two enzymes: MvaE, a dual function enzyme, acetoacetyl‐CoA thiolase and 3‐hydroxy‐3‐methylglutaryl‐CoA reductase of Enterococcus faecalis; MvaS, 3‐hydroxy‐3‐methylglutaryl‐CoA synthase of E. faecalis. MVA is produced by the heterologous MVA pathway from glycerol. (B) Enhanced cell growth and MVA production in P. putida KT2440. Both pST‐MVA and pSECRi(GlpR)‐Gen plasmids were co‐transformed into P. putida KT2440, and the transformants were cultured in M9 minimal medium containing 4 g l−1 glycerol in a 250‐ml baffled Erlenmeyer flask for 72 h at 30°C. Cell growth and mevalonate concentration were determined by spectrophotometer and HPLC respectively. As a control ((‐) CRISPRi), P. putida KT2440 harbouring the pSEVA221 plasmid was used. Each bar represents the mean value of the corresponding OD 600 or MVA concentration ± standard deviation of duplicate measurements from at least three independent experiments.
