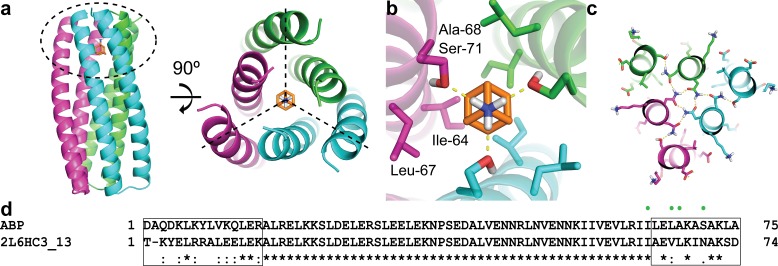
Figure 1. Computational design methodology.
(a) The homo-trimeric scaffold was designed to bind amantadine such that the C3 axes of the protein and the small molecule are aligned. Amantadine is colored orange and each monomer of ABP is colored magenta, green, or cyan. The amantadine binding site is highlighted by a dashed oval. (b) The binding pocket in ABP was designed to have polar serine residues (Ser-71) that hydrogen-bond (yellow dashed lines) to the amino group of amantadine and nonpolar residues (Ile-64, Leu-67, and Ala-68) to complement the shape of the hydrophobic moiety of amantadine. (c) The design model contains hydrogen-bond networks that specify the trimeric assembly of ABP. (d) A sequence alignment of ABP and 2L6HC3_13 is shown, with mutated regions shown in black boxes. Both sequences are preceded by a five-residue GHSMG pre-sequence (not shown) that result from the cloning strategy. The residues highlighted in (b) are annotated by green circles.