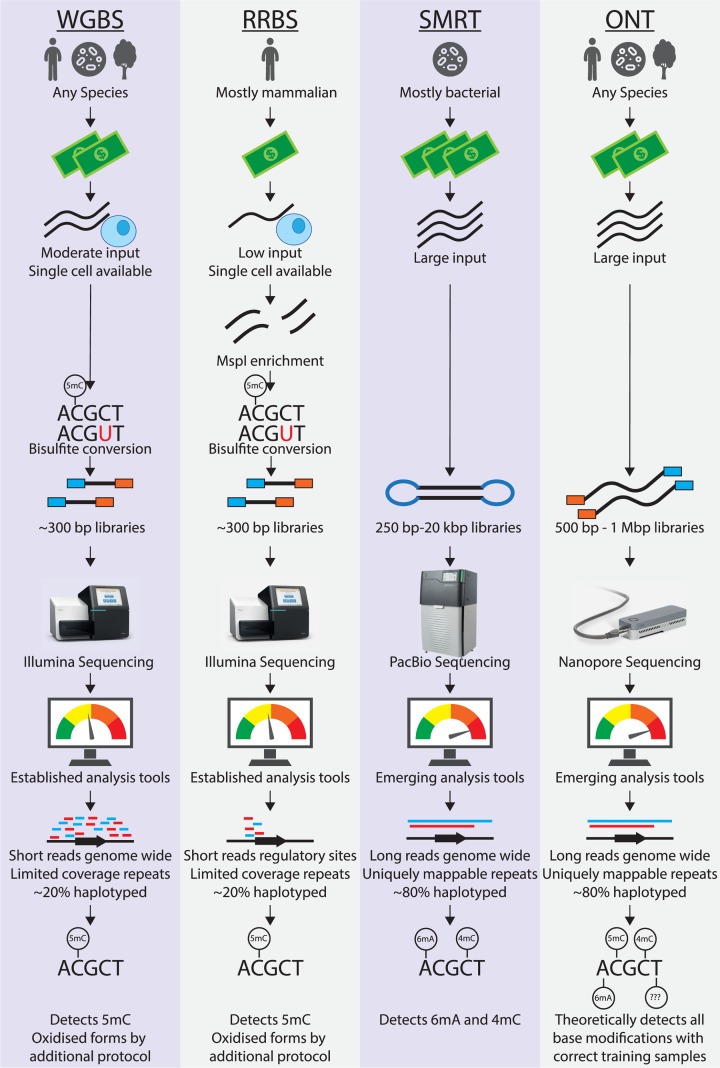
Figure 1. Detection of base modifications by bisulfite and long-read sequencing.
Whole-genome bisulfite sequencing (WGBS) provides accurate binary calls of cytosine methylation status at nucleotide resolution, but cannot distinguish between 5mC and 5hmC or detect other oxidised forms without additional techniques. The short reads have limitations in mapping to repeated sequences and haplotyping. Reduced representation bisulfite (RRBS) restricts the sequencing space to CpG islands. Both bisulfite protocols are compatible with single-cell genomics. PacBio single molecule real-time (SMRT) sequencing generates 250 b-20 kb reads with relatively low accuracy probabilistic calls for 4mC and 6mA modifications. Oxford Nanopore Technologies (ONT) nanopore sequencing produces 500 b-1 Mb reads with higher accuracy probabilistic calls for a range of modifications, including 5mC, 4mC, and 6mA. Long-read technologies offer a simpler library preparation workflow that avoids amplification biases, but they require more input material and more advanced data analysis. The longer read lengths improve mapping and haplotyping (estimates based on our experience with C57BL/6 x Cast/Eij F1 mice on Nanopore [62]).