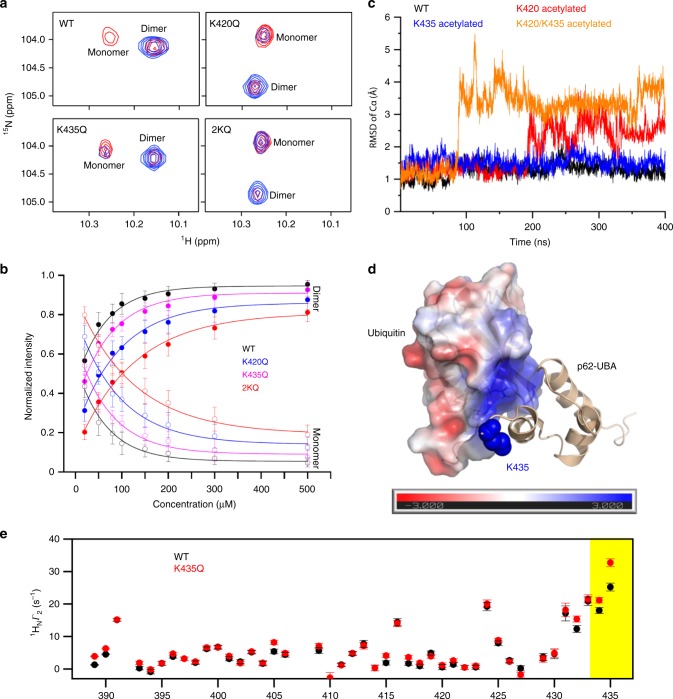
Fig. 6. Acetylation inhibits UBA dimerization and directly increases the affinity of p62 for ubiquitin.
a The W12 Hε1-Nε1 region of the 1H-15N HSQC spectra of 15N-labeled WT and acetylation-mimicking mutants of p62-UBA were measured at high (200 µM, blue) and low (20 µM, red) concentrations. b The dissociation constant of each of the indicated p62-UBA proteins was calculated based on the relative peak volumes for dimeric and monomeric species at different concentrations. The residues used for calculation are S399, L402, G405, S407, G411, W412, T419, and G425 Hε1-Nε1; the error bars indicate 1S.D. in the averaged relative peak volume at each protein concentration. c Representative trajectories of MD simulation for WT, K420 acetylated, K435 acetylated and K420/K435 acetylated p62-UBA dimers. Fluctuations in Cα RMSD of the dimer are plotted. d Structural model of the complex of p62-UBA and ubiquitin. The ubiquitin molecule is colored according to electrostatic potential, which is displayed on a scale from red (−3 kT/e) to blue (+3 kT/e). The side chain of K435 in p62-UBA is shown as blue spheres. e Paramagnetic relaxation enhancement for the transverse relaxation rate Γ2 of backbone amide protons for 15N-labeled WT or K435Q mutant of p62-UBA in complex with isotopically unlabeled ubiquitin carrying a nitroxide spin radical tag at the E24C site. The error bars indicate 1S.D. in the fitting of Γ2 rates. The yellow shaded area indicates the unstructured C-terminal tail of p62. Source data are provided as a Source Data file.