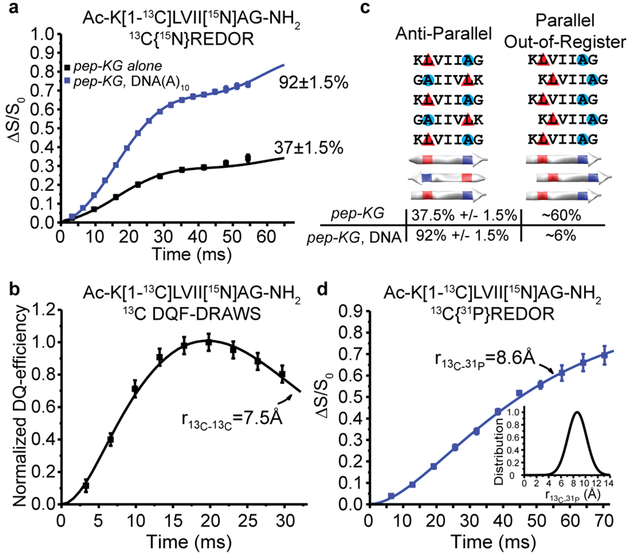
Figure 4. Structural characterization of DNA/Peptide co-assemblies.
(A) 13C{15N}REDOR measurements of 13C-15N distances for pep-KG 13C enriched at the leucine carbonyl ([1-13C]L) and 15N-enriched alanine ([15N]A) in both neat assemblies (black) and templated with DNA(A)10 (blue). Both assemblies have β-sheets with leucine H-bonded to alanine from adjacent strands indicating antiparallel, in-register β-strands with 37±1.5% of the peptides adopting this orientation in pep-KG assemblies, whereas 92±1.5% of the peptides in pep-KG/DNA(A)10 co-assemblies have this orientation. (B) 13C DQF-DRAWS measurement of 13C-13C distances in neat pep-KG assemblies assigned the ~60% of the [1-13C]Leu, not detected in the 13C{15N}REDOR experiment as parallel out-of-register by one amino acid β-sheets. (C) Models of peptide orientation and registry and their associated population from solid-state NMR measurements. Red triangles indicate position of [1-13C] and blue circles indicate 15N. (D) 13C{31P} REDOR of pep-KG/DNA(A)10 show that all of the [1-13C]L nuclei are proximal to at least one 31P nucleus and fit to a single distance of 8.6Å with a Gaussian distribution of 2.2Å (inset). Unless shown, error bars are the size of the data points and represent standard deviation.