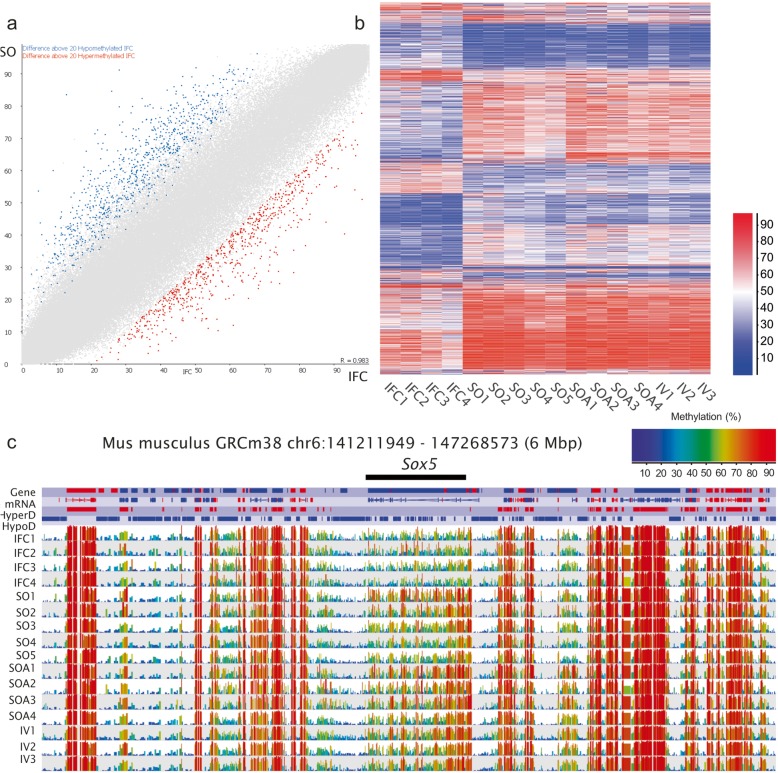
Fig. 3.
a Scatterplot for informative tiles (100 CpG window size, n = 195,170) in both IFC and SO. Data from replicates are pooled. Differentially methylated tiles (p < 0.05) identified by logistic regression and with a methylation difference of ≥ 20% are highlighted in blue or red (hypomethylated in IFC and hypermethylated in IFC, respectively). b Heat map after unsupervised hierarchical clustering of all differentially methylated tiles (p < 0.05, 100-CpG window size, n = 6362) between IFC and SO. The heatmap shows how biological replicates were consistent within groups and IFC differed in a similar way from SO, SOA, and IV for these differentially methylated sites. c SeqMonk screenshot of a 6 Mbp region of chromosome 6 showing methylation at the Sox5 locus, with 18 hypomethylated tiles in IFC. Each color-coded vertical bar in the screenshot represents the methylation value of a non-overlapping 100-CpG tile. Genes and oocyte mRNA are shown in red or blue depending on their direction of transcription (forward or reverse, respectively)