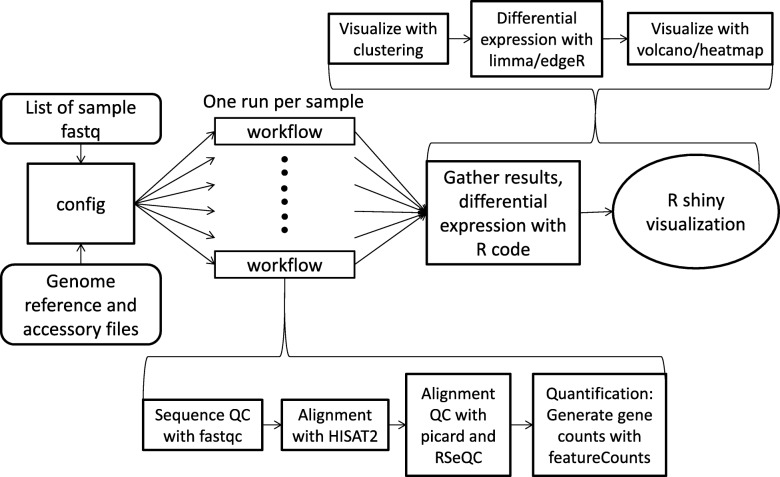
Fig. 1.
Schematic workflow of RNAseq pipeline. A list of fastq files and locations of necessary reference files are fed into config file which spawns a workflow run job for each sample. Results are gathered into an R data object and differential expression is calculated through provided R code. Visualization is provided through R shiny app