Figure 1.
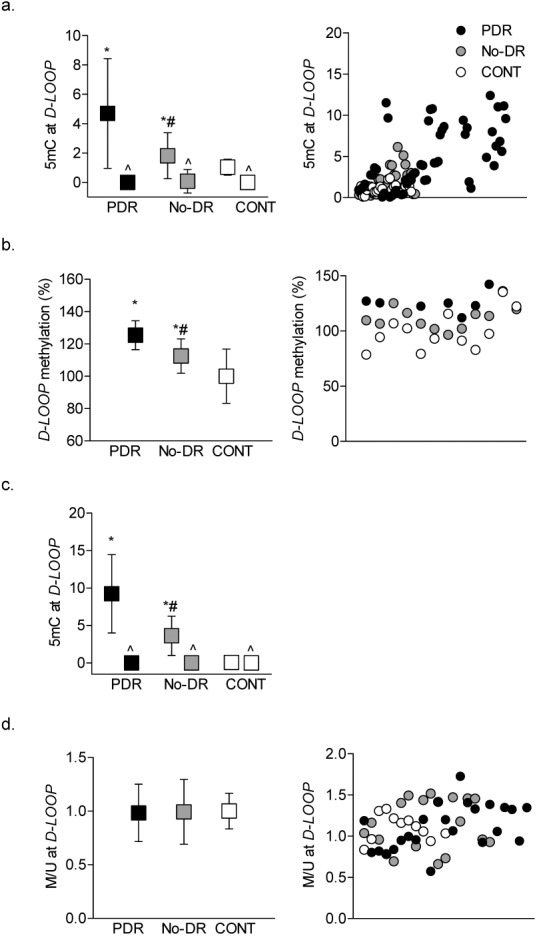
Methylation of mtDNA in the peripheral blood. Cytosine methylation in the D-Loop was quantified in the buffy coat (a) by a methylated DNA (MeDIP) immunoprecipitation kit. Fold change values obtained from qPCR were quantified by ddCt method and (b) by digesting DNA with methylation-sensitive HpaII endonuclease followed by qPCR using specific primers spanning the HpaII restriction site (-CCGG-). (c) The 5mC levels were quantified in purified mtDNA by MeDIP immunoprecipitation method. (d) Methylation-specific PCR was performed in the bisulfite-converted DNA, and the ratio of the methylated (M) to unmethylated (U) band was plotted. Value obtained from nondiabetic subjects was considered as one. The data are presented as mean ± SD obtained from 12 to 25 diabetic patients, each with proliferative retinopathy (PDR) or without retinopathy (No-DR), and nondiabetic control subjects (CONT). *,#P < 0.05 versus CONT and PDR groups respectively; ^IgG control antibody.
